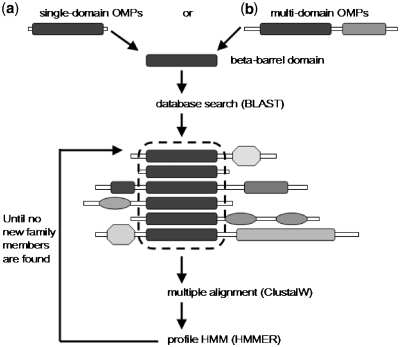
Figure 1.
Schematic ilustration of the procedure used to create the pHMMs that represent the TM domains of β-barrel OMPs. Either we were dealing with (a) single-domain OMPs or (b) multi-domain ones, following the literature we isolated the domain responsible for the β-barrel formation. Afterwards, we performed a BLAST search (47) against Uniprot using as query the β-barrel domain of the experimentally verified protein reported in the respective published reports, we created multiple alignments with the best-scoring results using ClustalW (48) and then, we built pHMMs for the particular domain using version 3.0b3 of the HMMER (49) software package. The pHMM was subsequently used to search for new family members and the procedure iterated until no new family members are found. For families that were represented in Pfam-B subset, we used the already deposited automatically generated multiple alignments.