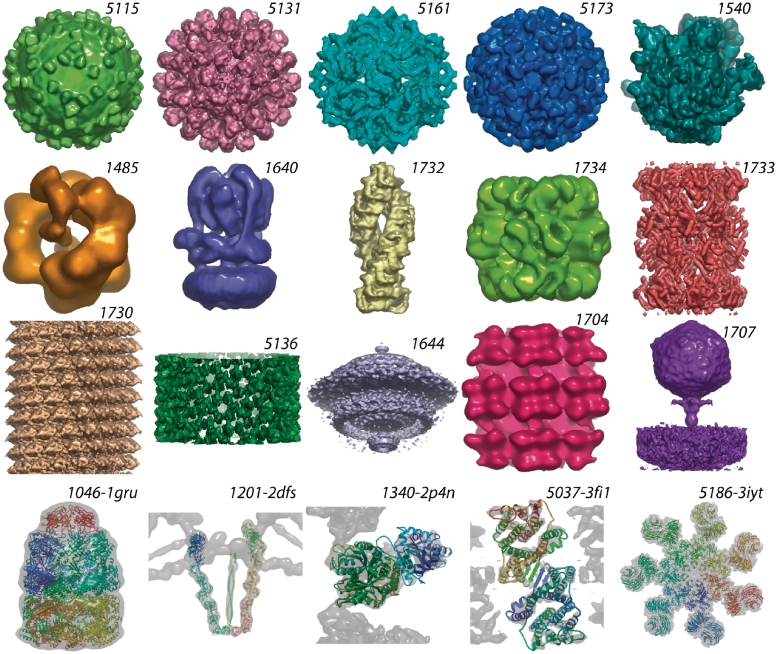
Figure 3.
Image gallery of EM structures in EMDB and PDB (labels are database ids with “EMD-” prefixes omitted for brevity; scaling is arbitrary). Icosahedral—5115: West Nile virus-antibody complex (53); 5131: wild-rabbit hemorrhagic disease virus (54); 5161: Partitivirus (55); 5173: Hepatitis E particles (56); other single particle: 1540: pre-translocational ribosome (57); 1485: DNA ligase–PCNA–DNA complex (58); 1640: yeast V-ATPase (59); 1732: tripeptidyl peptidase II (60); 1734: rubisco-methyltransferase complex (61); 1733: 20S proteasome (62); Helical—1730: tobacco mosaic virus (63); 5136: HIV-1 capsid tubular crystal (64); Tomogram averages—1644: Borrelia flagellar motor (65); 1704: Tula Hantavirus glycoprotein spikes (66); 1707: Podovirus P-SSP7 infecting Prochlorococcus marinus (67); Map+model pairs—1046-1gru: GroES–GroEL complex (68); 1201-2dfs: Myosin V (69); 1340-2p4n: kinesin bound to a microtubule (70); 5037-3fi1: ion-coupled transport protein NhaA (71); 5186-3iyt: apoptosome–procaspase-9 CARD complex (72). All images were created using OpenAstexViewer as implemented on the EMDB atlas pages for these entries.