Abstract
The National Institute of Agrobiological Sciences (NIAS) is implementing the NIAS Genebank Project for conservation and promotion of agrobiological genetic resources to contribute to the development and utilization of agriculture and agricultural products. The project’s databases (NIASGBdb; http://www.gene.affrc.go.jp/databases_en.php) consist of a genetic resource database and a plant diseases database, linked by a web retrieval database. The genetic resources database has plant and microorganism search systems to provide information on research materials, including passport and evaluation data for genetic resources with the desired properties. To facilitate genetic diversity research, several NIAS Core Collections have been developed. The NIAS Rice (Oryza sativa) Core Collection of Japanese Landraces contains information on simple sequence repeat (SSR) polymorphisms. SSR marker information for azuki bean (Vigna angularis) and black gram (V. mungo) and DNA sequence data from some selected Japanese strains of the genus Fusarium are also available. A database of plant diseases in Japan has been developed based on the listing of common names of plant diseases compiled by the Phytopathological Society of Japan. Relevant plant and microorganism genetic resources are associated with the plant disease names by the web retrieval database and can be obtained from the NIAS Genebank for research or educational purposes.
INTRODUCTION
The National Institute of Agrobiological Sciences (NIAS) Genebank (1) plays a leading role in the preservation and documentation of plant, microorganism and animal genetic resources related to food and agriculture in Japan (2). The NIAS Genebank manages this activity in collaboration with sub-banks located across Japan, with NIAS acting as the central bank. The NIAS Genebank has collected genetic resources in Japan and overseas. The collected genetic resources have been classified, evaluated, multiplied and preserved. Genetic resources in the public domain are distributed together with relevant information for breeding, scientific studies (including genome research) and educational purposes.
The genetic resources database has been built based on information collected during the exploration, evaluation, conservation and distribution management by the NIAS Genebank. The database consists of three categories: passport data, stock control data and characteristics/evaluation data. To manipulate these data, several data management programs have been developed. For the public data release, a web retrieval database has been developed with an external schema based on ANSI/SPARC three-level schema architecture (3); this was done to make the web retrieval applications independent of modifications of the genetic resources database schema. For the DNA Data Bank of Japan (DDBJ) (4), an efficient system was constructed using an object-oriented library as an external schema (5). For the NIAS Genebank, a denormalized database functions as an external schema. This schema is based on a relational database management system (RDBMS) so that applications can be flexibly developed by using various programming languages. The web retrieval database provides passport and evaluation data, which can be accessed through plant and microorganism search systems. Genotypic information is provided as a resource for genetic diversity research, as discussed in detail below.
The host-pathogen interaction is one of the most important interactions in biological research, and a central source for disease information such as the common name of the disease and the pathogen is helpful for researchers. Information on human and animal infectious diseases is provided in systems such as Gemina (6). For plant diseases, the American Phytopathological Society maintains a web page giving the common names of plant diseases (http://www.apsnet.org/online/common/). However, to study these plant diseases, it is necessary to have not only the background information but also a sample set of host plants and pathogens. The NIAS Genebank has developed a database of plant diseases in Japan, and it contains a function to link the plant diseases in the plant diseases database with plant and microorganism genetic resources in the genetic resources database.
DATABASE SYSTEMS FOR GENETIC RESOURCES
The data management system for genetic resources includes a genetic resources database and a web retrieval database.
Genetic resources database
The genetic resources database at the NIAS Genebank is based on a relational data model. The schema consists of about 500 tables, which can be classified into three categories: passport data, stock control data and characteristics/evaluation data. Passport data are the core of the genetic resources database. To identify individual genetic resources, accession numbers are assigned to every genetic resource when passport data are entered into the database. The accession number is a key descriptor linking the three data categories. JP and MAFF numbers denote the accession numbers of plants and microorganisms, respectively. The passport data contain the variety name, scientific name, origin, donor and registration date of each accession. The stock control data are used for Genebank management; they include sample location within the preservation unit and stock condition such as quantity and germination rate. The characteristics/evaluation data for plants include morphological characteristics and resistance to environment stress; for microorganisms, the data include information on pathogenicity and symbiosis. Information on the history and properties of registered genetic resources is added to the genetic resources database as the occasion demands.
Web retrieval database
The genetic resources database was designed to efficiently process the registration and update of the three categories of data. However, its normalized table structure is not necessarily suitable for the retrieval system, which must join relevant tables. To balance the requirements of the data storage system with the speed of retrieval, a denormalized web retrieval database was developed as the external schema. The web search systems are connected to the web retrieval database. In this way, the application programs of the search systems are not directly affected by the modification of schema in the genetic resource database. The web retrieval database has been constructed by joining many tables in the genetic resources database. The construction program for the web retrieval database automatically runs to obtain current data from the genetic resources database once per day.
Genetic resources search systems
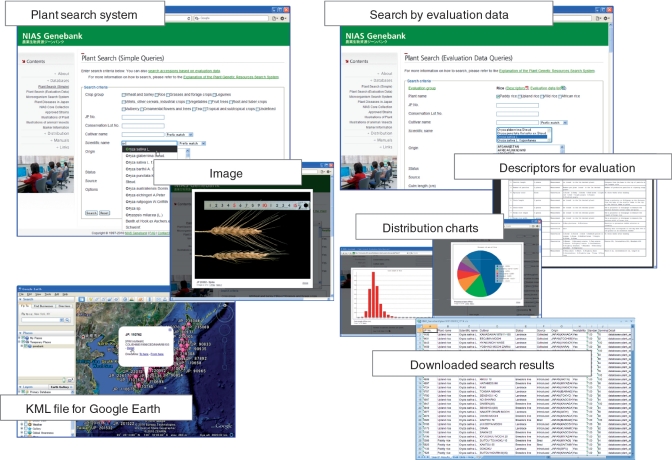
The plant search system for simple queries (http://www.gene.affrc.go.jp/databases-plant_search_en.php) has been developed in the web retrieval database to search passport data such as cultivar, origin and JP number (Figure 1). The auto-completer by Ajax suggests possible matches for entries typed into the scientific name and cultivar search fields. Searches for multiple selections (separated by commas) and partial match options are also available. The essential data for the matching entries are shown in tabular form, and more detailed information appears when the JP number is clicked. The search results can be downloaded as a Microsoft Excel file. A KML-formatted file can be downloaded for entries containing latitude and longitude data.
Figure 1.
Plant search system with various user-support options.
A plant search system for evaluation data queries (http://www.gene.affrc.go.jp/databases-plant_search_char_en.php) has also been developed. On the first page, the available crop groups are shown as a selection menu. When an evaluation crop group (for example, ‘rice’) is selected, a query form for the group is created based on the evaluation item name and data type, which are stored in the database as metadata (Figure 1). Minimum, maximum, average and median values are shown for numeric data and mode is shown for date data. To allow the user to understand the distribution of the evaluation data before data searching, the query form provides a function that draws the distribution shape as a circle or bar graph (Figure 1). A PDF file of the descriptors for characteristics and evaluation can be downloaded via a link in the evaluation data query form (Figure 1).
The microorganism search system (http://www.gene.affrc.go.jp/databases-micro_search_en.php) has been developed to search the passport data of registered microorganisms. The query form has the following categories: MAFF number, scientific name, designation, source, location and property. The search results appear in a tabular form containing information on the approved strain, reference strain and type strain.
RESOURCE INFORMATION FOR GENETIC DIVERSITY RESEARCH
The NIAS Genebank provides molecular information on genetic resources for use in genetic diversity research. This genotypic data is expected to contribute to a greater understanding of the association of genetic characteristics with phenotype.
NIAS core collection
A core collection is a limited set of accessions representing, with a minimum of repetitiveness, the genetic diversity of a crop species and its wild relatives (7). Adapting this concept to the NIAS Genebank, we developed the NIAS Core Collections (http://www.gene.affrc.go.jp/databases-core_collections_en.php). Each collection is designed for diversity research, and it consists of the minimum number of accessions necessary to represent the genetic diversity of the complete set of accessions maintained at the NIAS Genebank. The core collections are suitable for breeding, allele screening, linkage disequilibrium studies and crop evolution studies. The core collections currently available include global and Japanese cultivated rice and Japanese maize landraces.
The NIAS Global Rice Core Collection consists of 69 accessions. These accessions contain 90% of the RFLP alleles detected in about 300 accessions selected based on the passport data from the whole rice collection (about 30 000 accessions) maintained at the NIAS Genebank (8). The NIAS Rice Core Collection of Japanese Landraces consists of 50 accessions. These accessions contain 95% of the SSR alleles detected in about 240 accessions selected based on the passport data from the complete Japanese landrace collections (about 2000 accessions) maintained at the NIAS Genebank (9). The SSR polymorphism information on Japanese rice landraces is available for download in Excel format. The NIAS Maize Core Collection of Japanese Landraces consists of 17 Hokkaido and 69 other Japanese accessions (86 accessions in total). These were selected based on amplified fragment length polymorphism (AFLP) data from 300 Hokkaido landraces and 1000 other Japanese accessions, respectively, maintained at the NIAS Genebank.
Marker information
The NIAS Genebank has conserved the world’s most comprehensive and unique collections of cultivated and wild germplasm of Vigna, a member of the Leguminosae family. Among 98 species described in the genus Vigna, 9 domesticated species are presently cultivated in the world: azuki bean (V. angularis), mung bean (V. radiata), black gram (V. mungo), rice bean (V. umbellata), moth bean (V. aconitifolia), creole bean (V. reflexo-pilosa), cowpea (V. unguiculata), bambara groundnut (V. subterranea) and tuber cowpea (V. vexillata) (10). These species are important subsistence food crops because of their protein content, and they also contribute to soil fertility improvement via symbiotic nitrogen fixation. Representatives of all 9 cultivated species are currently maintained by the NIAS Genebank.
To facilitate the use of the genetic variation found in diverse germplasm, especially wild relatives, for crop improvement, the NIAS Genebank has been developing molecular markers and molecular linkage maps of Vigna crop species. SSR marker primer sequence information and marker positions on the linkage maps of azuki bean (11,12) and black gram (13) are currently available (http://www.gene.affrc.go.jp/databases-marker_information_en.php).
The azuki bean linkage map was constructed using 187 individuals of a BC1F1 population, and it contains primer sequence information for 196 SSR markers. The parental materials, cultivated azuki bean (V. angularis: JP81481) and wild azuki bean (V. nepalensis: JP107881), are available from the NIAS Genebank upon request.
The black gram linkage map was constructed using 180 individuals of a BC1F1 population derived from a cross between a mutant line (JP219132) selected for its large seed size from black gram cultivar ‘BC48’ (JP106710) and an accession ‘TC2210’ (JP107873) of its wild ancestor from India. The black gram map contains primer sequence information for 61 SSR markers.
Azuki bean linkage maps have been used to detect quantitative trait loci (QTLs) of domestication-related traits such as seed size, seed dormancy and pod shattering (14,15). Molecular genetic linkage maps for cowpea, mung bean and rice bean have already been developed, and SSR marker primer sequence information will soon be available from this site.
DNA sequence of approved fungal strains
Approved fungal strains for distribution are being selected based on microscopic inspection of their cultural characteristics and phylogenetic analyses of DNA sequences of gene regions suitable for strain identification.
Recently, molecular-phylogenetic studies of Fusarium and related species based on comparative analyses of DNA sequences have progressed significantly. Many circumscribed species within this genus have been found to be species complexes containing multiple species. Division or redefinition of these species is in progress. The NIAS Genebank has been performing DNA sequence analyses of its Japanese collection of Fusarium strains, analyzing sequences such as the histone H3 gene region, mitochondrion small-subunit ribosomal DNA (mtSSU rDNA), ribosomal DNA (rDNA), and internal transcribed spacer (ITS) regions (including partial sequences of 18S rDNA, ITS1, 5.8S rDNA, ITS2 and partial sequences of 28S rDNA), together with examining the cultural, morphological and phenotypic characteristics of these strains. These data are being used to establish a set of typical Japanese strains of Fusarium species based on the current taxonomic system (16). So far, 70 typical strains representing individual Fusarium species have been selected as ‘Approved Strains for Distribution (http://www.gene.affrc.go.jp/databases-micro_approved_en.php)’.
DATABASE OF PLANT DISEASES IN JAPAN
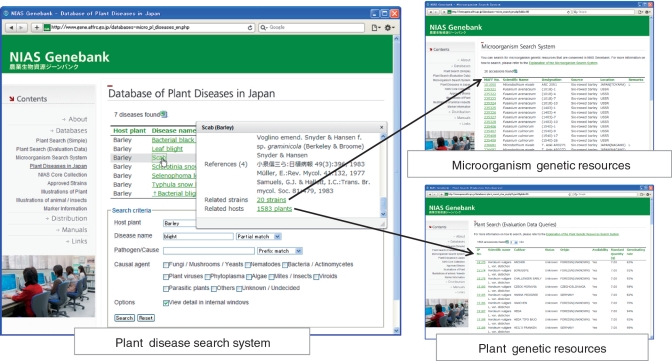
A database of plant diseases in Japan (http://www.gene.affrc.go.jp/databases-micro_pl_diseases_en.php) has been developed based on the book Common Names of Plant Diseases in Japan (17), compiled by the Phytopathological Society of Japan. In the database, 11 350 common names of diseases related to 1933 host plants and their pathogens are registered. The database contains host plant names, Japanese common names of diseases (converted to Roman alphabet), English disease names, scientific names of pathogens and other information from the book. These data are accessible via the web retrieval database (Figure 2). Although the original reference was written in Japanese, the plant disease search system is constructed for easy retrieval in English. Plant diseases can be retrieved by the search system according to host plant, disease name and pathogen/cause. Options such as auto-completion of entries, multiple selection using commas and partial matching are available for entries typed into the search fields. Search results are listed as a set containing plant name, disease name and pathogen(s). Detailed information appears when the disease name is clicked. These data and the URL for information on each disease can be downloaded in Excel format.
Figure 2.
Schematic diagram of relationships among NIAS Genebank databases and search systems.
Linkage of plant and microorganism genetic resources
One of the most important applications of this database is to search for host plants and plant pathogenic microbial strains in NIAS Genebank based on plant disease names. Matching plant and pathogen genetic resources have been selected using passport and evaluation data in the genetic resources database. The candidates for plant genetic resources are entries that have both the same scientific name as the host plant for a particular disease and a set of evaluation data for resistance to the disease. The candidates for microorganism genetic resources are entries that have the same scientific name as a known plant pathogen and have the plant hosts in the database listed as a source of the organism. The table of correspondence between plant diseases and the MAFF accession numbers of the candidate microorganisms is created in the web retrieval database and updated when the plant diseases database is modified. In the detailed information retrieved by the plant disease search system, the items ‘Related strains’ and ‘Related hosts’ are linked to microorganism and plant genetic resources, respectively. Taking the barley disease ‘scab’ as an example, 20 related strain accessions and 1583 related host plant accessions were retrieved (Figure 3).
Figure 3.
Plant disease database with links to plant and microorganism genetic resources.
FUTURE DIRECTIONS
DNA analysis of genetic resources stored at NIAS Genebank is continually being improved, so the DNA sequence data and marker information available for genetic diversity research is increasing. As new genotypic data become available, the information will be actively stored in NIASGBdb. We are developing additional core collections such as Japanese azuki bean landrace, amaranth and sorghum for distribution in the near future. In the plant diseases database, we are working to generate additional linkages with the genetic resources database by collecting additional microorganism genetic resources. Finally, information on biological interactions other than disease, such as symbiosis, will be established in the database.
FUNDING
This work was funded by National Institute of Agrobiological Sciences. Funding for open access charge: National Institute of Agrobiological Sciences.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the support of the Phytopathological Society of Japan for developing the plant diseases database. In particular, they thank the plant disease name committee of the society and the chairman, Dr Takao Tsukiboshi, for support in checking and modifying data in the database.
REFERENCES
- 1.Plucknett DL, Smith NJH, Williams JT, Anishetty NM. Gene Banks and the World’s Food. Princeton, NJ: Princeton University Press; 1987. [Google Scholar]
- 2.Okuno K, Shirata K, Niino T, Kawase M. Plant genetic resources in Japan: platforms and destinations to conserve and utilize plant genetic diversity. JARQ–Jpn. Agr. Res. Q. 2005;39:231–237. [Google Scholar]
- 3.Tsichritzis D, Klug A. The ANSI/X3/SPARC DBMS framework report of the study group on database management systems. Inform. Syst. 1978;3:173–191. [Google Scholar]
- 4.Kaminuma E, Mashima J, Kodama Y, Gojobori T, Ogasawara O, Okubo K, Takagi T, Nakamura Y. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 2010;38:D33–D38. doi: 10.1093/nar/gkp847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okayama T, Tamura T, Gojobori T, Tateno Y, Ikeo K, Miyazaki S, Fukami-Kobayashi K, Sugawara H. Formal design and implementation of an improved DDBJ DNA database with a new schema and object-oriented library. Bioinformatics. 1998;14:472–478. doi: 10.1093/bioinformatics/14.6.472. [DOI] [PubMed] [Google Scholar]
- 6.Schriml LM, Arze C, Nadendla S, Ganapathy A, Felix V, Mahurkar A, Phillippy K, Gussman A, Angiuoli S, Ghedin E, et al. GeMInA, Genomic Metadata for Infectious Agents, a geospatial surveillance pathogen database. Nucleic Acids Res. 2010;38:D754–D764. doi: 10.1093/nar/gkp832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel OH. Genetic perspectives of germplasm conservation. In: Arber W, Llimensee K, Peacock WJ, Starlinger P, editors. Genetic Manipulation: Impact on Man and Society. Cambridge: Cambridge University Press; 1984. pp. 161–170. [Google Scholar]
- 8.Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M. Development of an RFLP-based rice diversity research set of germplasm. Breeding Sci. 2005;55:431–440. [Google Scholar]
- 9.Ebana K, Kojima Y, Fukuoka S, Nagamine T, Kawase M. Development of mini core collection of Japanese rice landrace. Breeding Sci. 2008;58:281–291. [Google Scholar]
- 10.Tomooka N, Vaughan DA, Maxted N, Moss H. The Asian Vigna. Genus Vigna subgenus Ceratotropis genetic resources. Kluwer Academic Publishers, Dordrecht, The Netherlands. 2002 [Google Scholar]
- 11.Wang XW, Kaga A, Tomooka N, Vaughan DA. The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi] Theor. Appl. Genet. 2004;109:352–360. doi: 10.1007/s00122-004-1634-8. [DOI] [PubMed] [Google Scholar]
- 12.Han OK, Kaga A, Isemura T, Wang XW, Tomooka N, Vaughan DA. A genetic linkage map for azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi] Theor. Appl. Genet. 2005;111:1278–1287. doi: 10.1007/s00122-005-0046-8. [DOI] [PubMed] [Google Scholar]
- 13.Chaitieng B, Kaga A, Tomooka N, Isemura T, Kuroda Y, Vaughan DA. Development of a black gram [Vigna mungo (L.) Hepper] linkage map and its comparison with an azuki bean [Vigna angularis (Willd.) Ohwi and Ohashi] linkage map. Theor. Appl. Genet. 2006;113:1261–1269. doi: 10.1007/s00122-006-0380-5. [DOI] [PubMed] [Google Scholar]
- 14.Isemura T, Kaga A, Konishi S, Ando T, Tomooka N, Han O, Vaughan DA. Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm season legumes. Ann. Bot. 2007;100:1053–1071. doi: 10.1093/aob/mcm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaga A, Isemura T, Tomooka N, Vaughan DA. The genetics of domestication of the azuki bean (Vigna angularis) Genetics. 2008;178:1013–1036. doi: 10.1534/genetics.107.078451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki T. Taxonomic System of the genus. Fusarium. Microbiol. Cult. Coll. 2009;25:1–12. (in Japanese) [Google Scholar]
- 17.The Phytopathological Society of Japan. Common Names of Plant Diseases in Japan: First edition. Tokyo, Japan: Japan Plant Protection Association; 2000. (in Japanese) [Google Scholar]