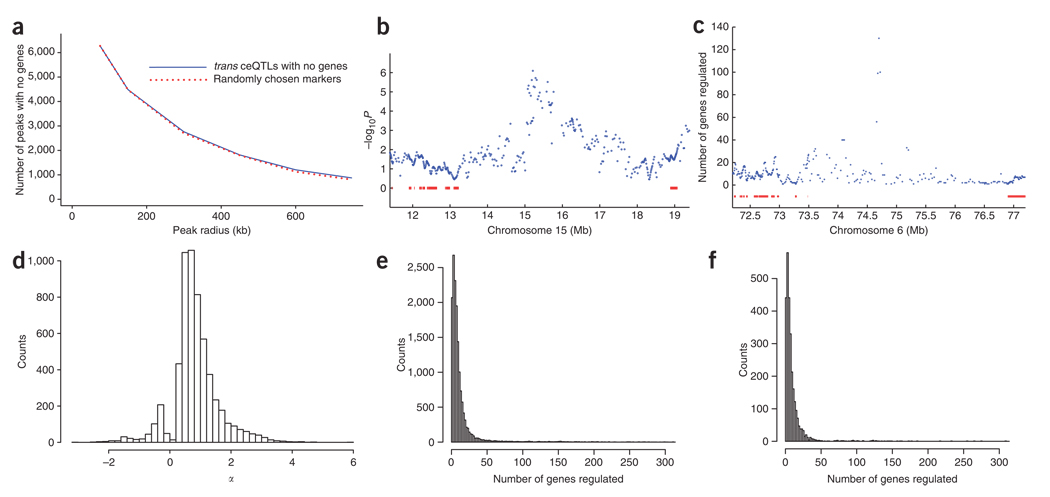
Figure 7.
Trans ceQTLs lacking known genes. (a) Relationship between number of trans ceQTLs with no genes and radius from peak marker (solid line). Trans ceQTLs regulating >1 gene are counted once only. The same relationship for randomly chosen markers is also shown (broken line). (b) Example of trans ceQTL on chromosome 15 at 15.3 Mb with no known genes underneath regulating the Maoa (monoamine oxidase A) gene on the X chromosome. Known genes are shown as red bars, (c) Example of a trans ceQTL hotspot with no known genes underneath. This trans ceQTL hotspot, located at 74.75 Mb on chromosome 6, regulates 130 genes. (d) Counts of trans loci with no genes and FDR < 0.4 versus α. Trans ceQTLs lacking genes had smaller mean α values than those with genes (0.832 ± 0.01 and 0.986 ± 0.006, respectively, permutation t-test, P < 2 × 10−5; see also Figure 6b). (e) Counts of trans ceQTLs with FDR < 0.4 versus numbers of regulated genes. (f) Counts of trans ceQTLs with no genes and FDR < 0.4 versus numbers of regulated genes. Fewer mean numbers of genes were regulated by trans ceQTLs lacking genes than by trans ceQTLs with genes (11.35 ± 0.48 and 13.49 ± 0.21 genes, respectively, permutation t-test, P < 2 × 10−5).