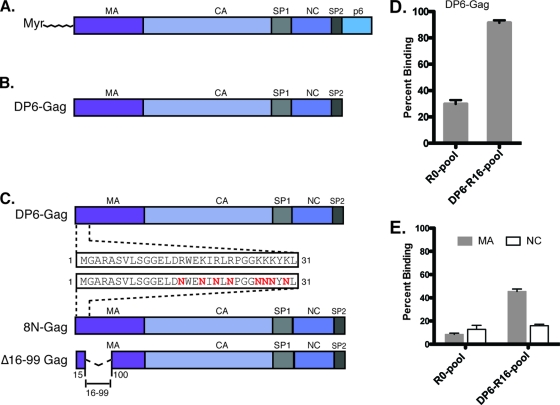
FIG. 1.
Diagrammatic representation of the various proteins used for RNA selections and aptamer analysis. (A) Wild-type HIV-1 Gag polyprotein. (B) The DP6-Gag protein used for selections lack the N-terminal myristate and the late domain p6. (C) 8N-Gag with eight basic amino acid residues of MA mutated to asparagines and Δ16-99-Gag, with a deletion in between amino acid residues 16 and 99. (D) The binding of RNAs from the R0 and R16 pools to DP6-Gag. (E) The binding of RNAs from the R0 and R16 pools to nonmyristoylated matrix (gray bars) and nucleocapsid (open bars). All binding reactions were performed with 10 nM aptamer RNAs and 1 μM protein, in the presence of a 5-fold molar excess of yeast tRNA to eliminate nonspecific binding.