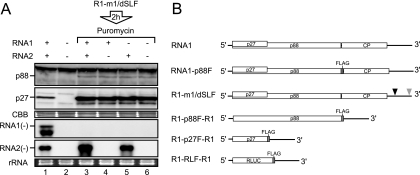
FIG. 1.
RNA2, but not RNA1, served as a template for negative-strand RNA synthesis in the presence of a translation inhibitor. (A) RNA1 and RNA2 (30 nM each) were incubated in BYL at 17°C for 4 h (lane 1). Capped R1-m1/dSLF (30 nM) was incubated in BYL for the production of replicase proteins. After 2 h of incubation at 17°C, puromycin was added into the lysate at a concentration of 100 μg/ml. Subsequently, RNA1 and/or RNA2 (30 nM each) was added to the lysates and incubated at 17°C for 2 h (lanes 3 to 5). Total RNA and protein were extracted and used for Northern and Western blot analyses, respectively. Western blotting was performed using an anti-p27 antiserum. Northern blotting was performed using digoxigenin-labeled RNA probes that were complementary to full-length negative-strand RNA1 and RNA2. RNA1(−) and RNA2(−) indicate negative-strand RNA1 and RNA2, respectively. Coomassie brilliant blue (CBB)-stained cellular proteins are shown below the Western blots, as loading controls. EtBr-stained rRNAs are shown below the Northern blots, as loading controls. (B) Schematic representation of the RCNMV RNA1 mutants used in this study. Open boxes denote the open reading frames of replicase proteins (p27 and p88), coat protein (CP), and Renilla luciferase (R-Luc). The bold lines in the 5′ and 3′ proximal regions denote the 5′ and 3′ untranslated regions (UTRs) of RNA1, respectively. FLAG tag sequences are represented by gray boxes. Black and gray triangles indicate deleted regions in the 3′ UTR of RNA1. These deletions, denoted by the black and gray triangles, abrogate the generation of SR1f and the synthesis of negative-strand RNA1, respectively (18, 19).