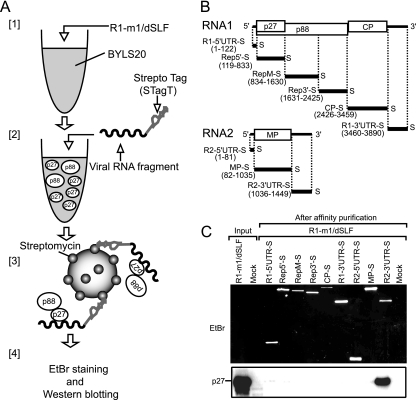
FIG. 2.
p27 was purified efficiently with the 3′ UTR of RNA2 by using Strepto Tag affinity purification. (A) Depiction of the purification steps of replicase proteins by using the Strepto Tag method. [1] Viral replicase proteins were expressed from R1-m1/dSLF (30 nM) in the 20,000 × g supernatant fraction of an evacuolated tobacco BY-2 protoplast lysate (BYLS20). [2] Various viral RNA fragments that were fused to the Strepto Tag sequence at the 3′ ends were incubated in the lysate. [3] These lysates were applied to affinity columns containing streptomycin immobilized to Sepharose. [4] After washing, trapped probe RNA-protein complexes were eluted with the antibiotic. Finally, probe RNAs and viral replicase proteins were visualized and detected using EtBr staining and Western blotting, respectively. (B) Schematic representation of the RCNMV genomic RNAs and Strepto Tag-fused viral RNA fragments used in this assay. Bold lines indicate the virus-derived sequence of Strepto Tag-fused viral RNA fragments, with the nucleotide numbers indicated at the 5′ and 3′ ends. The “S” at the 3′ end of viral RNA fragments indicates the Strepto Tag sequence. (C) p27 was purified efficiently with the 3′ UTR of RNA2 by using the Strepto Tag affinity purification scheme outlined in panel A. The input lane contained 1.6% of the extract used for the Strepto Tag affinity purification. Purified RNAs (top) were visualized by EtBr fluorescence. Purified p27 (bottom) was detected by Western blotting using an anti-p27 antiserum.