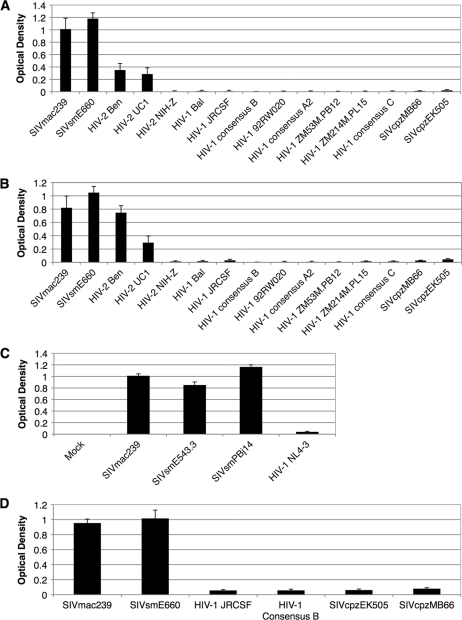
FIG. 10.
The jacalin and PNA binding capacities of SIVmac/sm, HIV-2 Ben, and HIV-2 UC1 gp120s differ from those of HIV-2 NIH-Z, HIV-1, and SIVcpz gp120 proteins. (A) Jacalin binding capacities of SIVmac, SIVsm, HIV-2, HIV-1, and SIVcpz gp120 proteins. Equivalent amounts of histidine-tagged gp120 proteins were each loaded into six wells of a high-protein-binding plate. Jacalin-HRP was used to probe each sample in quadruplicate. A histidine-HRP antibody was used to probe each sample in duplicate. The mean jacalin signal was normalized to the mean signal from the histidine tag. The normalized jacalin signal (± standard deviation) is shown. (B) Same as panel A but without normalization. (C) Comparison of the jacalin binding capacities of SIVmac, SIVsm, and HIV-1 gp120s. HEK293T cells were transiently transfected either with no DNA (mock) or with a gp120 eukaryotic expression vector for SIVmac, SIVsm, or HIV-1. Env was purified from the cell culture supernatant with lectin from Galanthus nivalis. Equivalent amounts of total protein were loaded into the wells of a high-protein-binding plate. Jacalin-HRP was used to probe each sample in quadruplicate. The mean jacalin signal (± standard deviation) is shown. (D) Lectin from peanut (PNA) binds to the gp120 of SIVmac/sm but not to the gp120 of SIVcpz or HIV-1. Equivalent amounts of histidine-tagged gp120 proteins were each loaded into six wells of a high-protein-binding plate. PNA-HRP was used to probe each sample in quadruplicate. A histidine-HRP antibody was used to probe each sample in duplicate. The mean PNA signal was normalized to the mean signal from the histidine tag. The normalized PNA signal (± standard deviation) is shown.