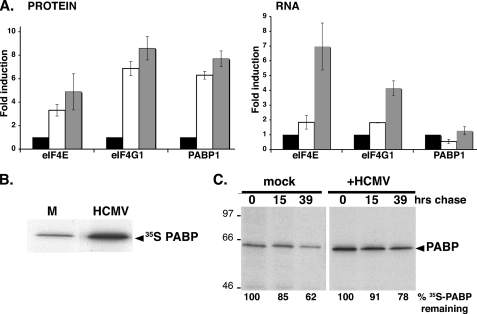
FIG. 3.
Translational control of PABP levels in HCMV-infected cells. (A) Analysis of eIF4E, eIF4G, and PABP mRNA abundance in HCMV-infected cells. Serum-starved NHDFs were mock infected (0 h) or infected with HCMV (MOI = 5). At 0 hpi (black bars), 70 hpi (white bars), and 96 hpi (gray bars), total protein (left) was fractionated by SDS-PAGE and analyzed by immunoblotting with the indicated antisera. Chemiluminescence images were captured and quantified with a Bio-Rad ChemiDoc XR5 system. Total RNA (right) was isolated, and eIF4G1, eIF4E, and PABP mRNA levels were determined by real-time RT-PCR (normalized to actin). (B) Stimulation of new PABP synthesis in HCMV-infected cells. NHDFs were infected as described for panel A. At 24 hpi, cultures were pulse-labeled for 2 h with [35S]Met-Cys, and PABP was immunoprecipitated. Immune complexes were fractionated by SDS-PAGE and visualized by autoradiography. (C) PABP stability is similar in both mock- and HCMV-infected cells. NHDFs infected as described for panel A were pulse-labeled at 50 hpi as described for panel B and then returned to unlabeled medium for the indicated chase times. Total protein was subsequently isolated, and PABP was immunoprecipitated and analyzed as described for panel B. The mobility of molecular mass standards (in kilodaltons) is shown to the left. The percentage of [35S]PABP remaining over time was quantified by densitometry. In each case (mock versus HCMV infection), the amount of radiolabeled PABP after the pulse (0 h chase) was set at 100%.