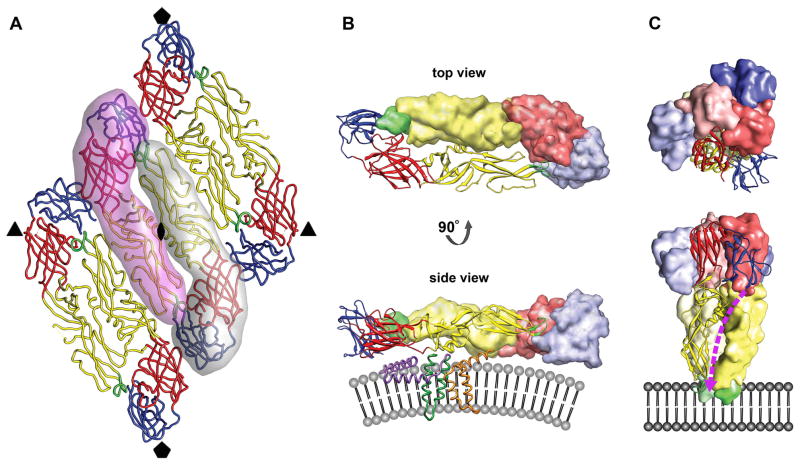
Fig. 2.
The oligomeric structure of the E glycoprotein. (A) Raft organization of three E homodimers as observed for mature flavivirions. The relative positions of the neighboring icosahedral symmetry axes are indicated by symbols. The elongated E ectodomain has three distinct, structurally defined domains that are joined by flexible hinges. DI, DII, and DIII are colored red, yellow, and blue, respectively. The fusion loop is shown in green. Two E monomers forming the central homodimer at the icosahedral twofold axis are emphasized by shadows. (B) The prefusion head-to-tail homodimer arrangement of the E glycoprotein shown from the top (looking towards the center of the virus) and from the side after a 90° rotation around its long axis. One monomer is rendered as a ribbon diagram, showing its three domains largely adopting β-sheet folds. The other monomer is represented as a surface shaded volume. The fusion loop of one E protein is buried in a hydrophobic pocket at the DI–DIII interface of the other E molecule. The side view shows that each E monomer is structurally divided into the ectodomain that elongates parallel to the viral lipid envelope and a two-helix transmembrane anchor (dark green). The α-helical stem region (purple), half-buried in the outer lipid leaflet, connects the C-terminus of the E ectodomain with the transmembrane helices. The M protein (orange) also contains two antiparallel transmembrane helices and is essentially buried under the E protein layer. The stem-anchor region is only shown for the E monomer depicted as a ribbon diagram. Similarly, only one M protein is illustrated. (C) The postfusion homotrimer arrangement of the E glycoprotein shown from the top and from the side after a 90° rotation. The E monomers are oriented parallel to one another with their fusion peptides exposed on one end of the trimer. DIII of E has undergone a major rotational displacement to the side of DI and towards DII. The stem region (dashed purple line) is predicted to bind in a hydrophobic groove that extends along the interface of two neighboring DIIs of the E trimer towards the fusion loop. The irreversible fold-back conversion of the E protein into the post-fusion hairpin-like structure and zipping up of the stem brings the E transmembrane helices (connected via the stem to DIII of the ectodomain) in juxtaposition with the membrane-embedded fusion loop at the tip of DII.