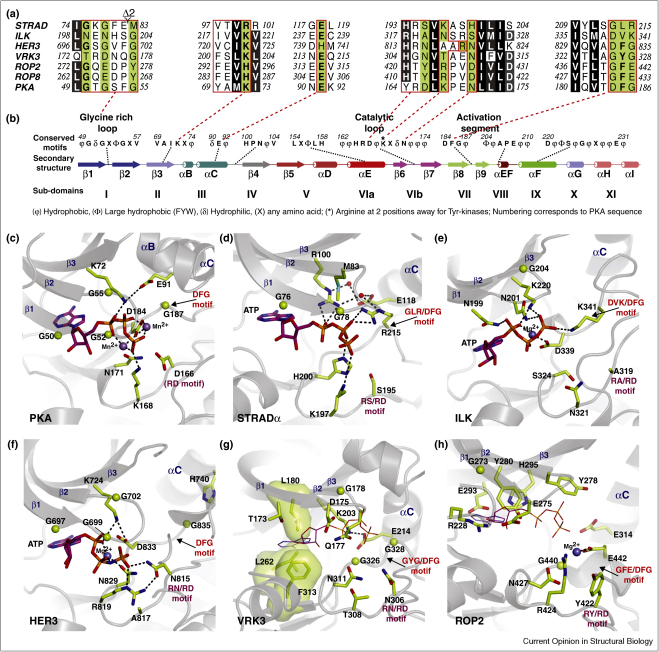
Figure 1.
Degradation of pseudokinase nucleotide binding pockets and ‘active’ sites. (a) Multiple sequence alignment of pseudokinases and the canonical protein kinase PKA. Highlighted in green are key residues that are deemed to be essential for activity in eukaryotic protein kinases. (b) Kinase domain secondary structure, subdomains and conservation of key motifs. The secondary structure is labelled and the consensus sequence of common motifs and key conserved loops are given. These were deduced from multiple sequence alignments of representative protein kinases from each branch of the human kinome [28••] and the study of Kannan et al., 2007 [9]. The subdomains are labelled using the nomenclature described by Hanks et al., 1988 [3] and Taylor and Radzio-Andzelm, 1994 [4]. (c–h) Nucleotide binding pockets and active sites of PKA (PDBID 1ATP [1], STRADα (PDBID 3GNI [28••], ILK (PDBID 3KMW; [36••], HER3 (PDBID 3KEX; [39•], VRK3 (PDBID 2JII; [40••]) and ROP2 (PDBID 2W1Z; [44••]). ATP is shown as sticks with magenta carbon atoms. For VRK3 and ROP2, ATP (shown as lines) bound to PKA was modelled in the VRK3 and ROP2 structures by superposition of the PKA structure (PDBID 1ATP). Glycine residues are depicted as green spheres, water molecules are shown as red spheres, Mn2+ atoms as purple spheres and Mg2+ as blue spheres. Hydrogen bonding interactions are represented by dashed lines, and residues making up the hydrophobic spine of VRK3 are shown as sticks and transparent surface.