Fig. 2.
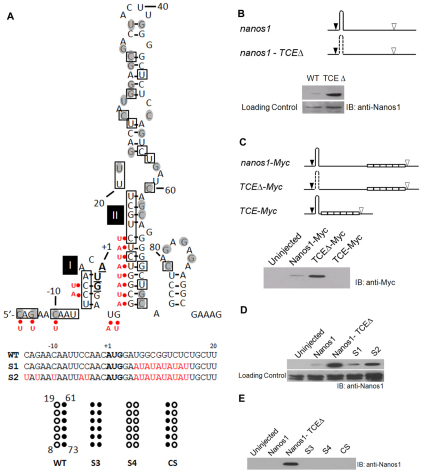
Identification of the nanos1 translational control element (TCE). (A) Summary of structural probing of the 5′ terminal region of nanos1 transcript with RNases A, T1 and V1. Secondary structure of the 5′ terminal region of nanos1 (–15 to +95) is shown as predicted by MFOLD (Zuker, 2003). The start codon nucleotides are in bold and underlined. The nucleotides sensitive to single-strand-specific RNase are within shaded circles and those sensitive to base-paired-specific RNase are in open boxes. AU substitutions are indicated by red dots. The S3, S4 and CS mutants are illustrated beneath, alongside the wild-type (WT) sequence. (B) Deletion of the TCE relieves repression. Translation of wild-type nanos1 and a TCE deletion mutant (TCEΔ) injected into Xenopus oocytes. The predicted nanos1 TCE (+8 to +73) is shown as a hairpin structure. Start (black arrowhead) and stop (white arrowhead) codons are indicated; dashed line signifies deletion. Immunoblot (IB) with anti-Nanos1 antibody. (C) The TCE is both necessary and sufficient for nanos1 repression. nanos1-Myc and deletion mutants, TCEΔ-Myc and TCE-Myc, were injected into oocytes and their translation analyzed by blotting with anti-Myc antibody. Myc tags are shown as open white. (D) Disruption of base-pairs in Stem-loops I and II promotes translation. Translation in oocytes of mutants S1 and S2, analyzed as detailed in B. (E) Substitution (S3, S4) or compensatory (CS) changes failed to relieve repression. See also Figs S2 and S3 in the supplementary material.