Abstract
A central challenge of synthetic biology is to enable the growth of living systems using parts that are not derived from nature, but designed and synthesized in the laboratory. As an initial step toward achieving this goal, we probed the ability of a collection of >106 de novo designed proteins to provide biological functions necessary to sustain cell growth. Our collection of proteins was drawn from a combinatorial library of 102-residue sequences, designed by binary patterning of polar and nonpolar residues to fold into stable 4-helix bundles. We probed the capacity of proteins from this library to function in vivo by testing their abilities to rescue 27 different knockout strains of Escherichia coli, each deleted for a conditionally essential gene. Four different strains – ΔserB, ΔgltA, ΔilvA, and Δfes – were rescued by specific sequences from our library. Further experiments demonstrated that a strain simultaneously deleted for all four genes was rescued by co-expression of four novel sequences. Thus, cells deleted for ∼0.1% of the E. coli genome (and ∼1% of the genes required for growth under nutrient-poor conditions) can be sustained by sequences designed de novo.
Introduction
In 1906 Jacques Loeb suggested that the synthesis of life is a significant goal of biology [1]. A century later, the construction of natural genomes from off-the-shelf chemicals [2], [3] demonstrates significant progress toward achieving this goal. Until now, however, most advances in synthetic biology have relied on collections of parts – genes, proteins, and regulatory elements – derived from sequences that already exist in nature. Must the toolkit of life be so restricted? Natural sequences comprise only a miniscule fraction of the theoretical sequence space that is possible for genes and proteins. Indeed, a collection containing just a single molecule of every one of the 20100 possible 100-residue proteins would fill a volume larger than a mole of universes [4]. From this enormous potential for diversity, natural selection has operated over billions of years to yield a relatively small collection of sequences: Life is sustained by only ∼4,000 genes in E. coli and approximately 5-fold more in humans [5], [6]. These considerations might lead to the supposition that genes and proteins capable of sustaining life are somehow “special.” Is this true? Or might we find functional molecular parts in a collection of artificial sequences designed “from scratch” in the laboratory?
To address these questions, we probed the ability of unevolved sequence space to encode functions that enable cell growth. We designed and constructed a collection of artificial genes encoding approximately 1.5×106 novel amino acid sequences. Because folding into a stable 3-dimensional structure is a prerequisite for most biological functions, we did not construct this collection of proteins from random sequences. Instead, we used the binary code strategy for protein design, shown previously to facilitate the production of large combinatorial libraries of folded proteins [7], [8]. In brief, the binary code strategy posits that stably folded proteins can be encoded by specifying the sequence pattern of polar and nonpolar residues (the binary pattern) to coincide with the exposed and buried parts of a structure, respectively. For example, to design a helical bundle comprising α-helices with hydrophilic faces exposed to aqueous solvent and hydrophobic faces buried in the interior of a protein, the binary pattern of polar (P) and nonpolar (N) residues should be PNPPNNPPNPPNNP, consistent with the structural repeat of 3.6 residues per alpha-helical turn [9], [10]. For the current studies, we used the binary code strategy to design and construct a library of sequences designed to fold into 102-residue 4-helix bundles. Our strategies for protein design and selecting biological function are summarized in Figure 1. Details describing the construction and biophysical characterization of the library are presented elsewhere [11], [12].
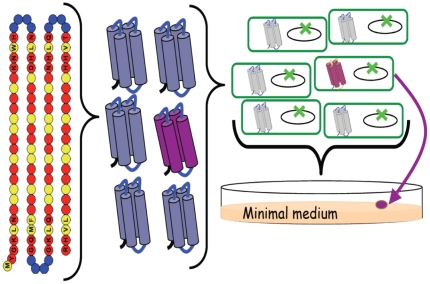
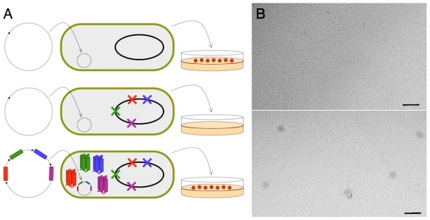
Figure 1. Design of a collection of novel proteins and rescue of E. coli auxotrophs.
Binary pattern of polar (red) and nonpolar (yellow) residues designed to fold into 4-helix bundles (7, 8). Library construction is facilitated by the organization of the genetic code: Six polar residues (Lys, His, Glu, Gln, Asp, and Asn) are encoded by the degenerate codon VAN, and five nonpolar residues (Met, Leu, Ile, Val, and Phe) are encoded by the degenerate codon NTN. Combinatorial turn positions are encoded by the degenerate codon VRS, which encodes Gln, Glu, Asn, Asp, His, Lys, Arg, Ser, and Gly. (V = A, G, or C; R = A or G; N = A, G, C, or T). Circles with letters indicate fixed residues, and empty circles indicate combinatorially diverse positions. The theoretical diversity of this library is 522×634×812 = 5×1052. The substantially smaller experimental library (1.5×106 synthetic genes cloned on a high expression plasmid) was transformed into a strain of E. coli in which an endogenous gene had been deleted (green X). Colony formation on minimal media indicates a novel sequence (purple) enables cell growth under selective conditions.
Here we demonstrate that novel proteins from this binary patterned library can rescue E. coli cells that lack certain natural proteins required for cell viability. The novel proteins are substantially less active, and may function by different mechanisms than the natural proteins they replace. Nonetheless, co-expression of several novel proteins rescues a strain in which multiple natural genes were deleted simultaneously. These findings show that an organism, which would otherwise not grow, can be sustained by macromolecules devised in the laboratory.
Results and Discussion
De novo designed sequences rescue single-gene knockout strains of E. coli
Previous work in our laboratory showed that several purified proteins from our binary patterned alpha-helical libraries can bind a cofactor and perform enzyme-like functions, including peroxidase, lipase, and esterase activities [13]. These findings led us to question whether proteins from this library might also provide biological functions that enable cell growth. To address this question, we tested the ability of binary patterned proteins to rescue strains of E. coli in which a conditionally essential function had been deleted. These strains were obtained from the Keio collection, which contains all the viable single-gene knockout strains of E. coli [5]. We tested 27 auxotrophs that grow on rich media, but fail to grow on minimal media (see legend for Table 1). Each auxotroph was transformed with a library of synthetic genes carried on the expression vector pCA24NMAF2 [14]. Typically, we obtained 5–10 million transformants, thereby ensuring reasonably deep coverage of the library of 1.5×106 de novo sequences. As negative controls, each auxotroph was transformed with the empty vector or the same vector expressing beta-galactosidase. Transformed cells were spread either on LB (rich) or on M9-glucose (minimal) media supplemented with isopropyl-beta-D-thiogalactoside (IPTG) to induce expression, and the formation of colonies was monitored (Figures 1 & 2).
Table 1. Complementation frequency and time required for colony formation on selective media (M9-glucose).
| Complementation Frequency | Time to Grow (Days) | |
| ΔserB | ∼1/50,000 | 4 |
| ΔgltA | ∼1/1,000,000 | 4 |
| ΔilvA | ∼1/3,000 | 3 |
| Δfes | ∼1/10,000 | 2 |
Novel proteins capable of enabling cell growth were isolated for 4 strains. Complementation frequency was calculated by dividing the number of colonies on selective plates (rescued transformants) by the number of colonies on rich plates (total transformants). This apparent frequency was then adjusted to estimate the true complementation rate by correcting for any false positives that failed to restreak or retransform. The following additional strains from the Keio collection were tested, but were not rescued by sequences from the library: ΔargB, ΔargE, ΔargH, ΔaroB, ΔaroC, ΔaroD, ΔaroE, ΔbioA, ΔcysG, ΔglnA, ΔhisA, ΔilvC, ΔilvD, ΔleuB, ΔleuC, ΔleuD, ΔpdxA, Δppc, ΔproB, ΔpurF, ΔpyrF, ΔthrB, and ΔthrC.
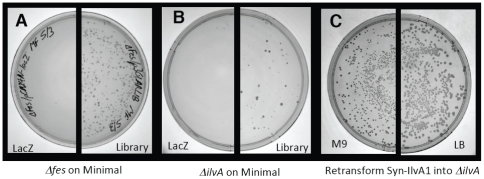
Figure 2. Rescue of E. coli auxotrophs by de novo proteins.
(A) E. coli strain BW25113 Δfes was transformed with a control plasmid overexpressing LacZ (left half), or with a library of plasmids encoding the collection of novel proteins (right half). Cells were plated on M9-glucose supplemented with 50 µM IPTG, 30 µg ml−1 kanamycin, and 34 µg ml-1 chloramphenicol. Cells transformed with the control plasmid fail to grow, whereas cells transformed with the library yield occasional colonies. (B) Similar results for ΔilvA. (Similar results were also observed for ΔserB and ΔgltA – data not shown.) (C) Re-transformation of ΔilvA cells with a single sequence, syn-ilvA-1. A similar number of colonies grow on M9-glucose (left) as on LB (right). Results for other sequences were similar to those shown in (C), and are summarized in Figure S1.
On rich media, all transformed cells grew, regardless of whether they received the control lacZ gene or a gene encoding a novel protein. As expected, on minimal media, auxotrophs transformed with the control plasmids failed to grow. (If the negative controls grew, the strain was considered ‘leaky’ and not used for further studies.) In most cases, auxotrophs transformed with the collection of novel sequences also failed to produce colonies, even after weeks of incubation. This indicates that proteins from the collection could not rescue these strains. However, for four auxotrophs – ΔserB, ΔgltA, ΔilvA, and Δfes – colonies formed on minimal plates after several days of incubation (Figures 2A & 2B). Growth of these cells under selective conditions suggests that a novel gene carried by the plasmid complements the deletion. The rates of complementation and the times required for colony formation are summarized in Table 1.
To confirm that the colonies observed on selective media resulted from the uptake of a novel gene and not from an adaptive mutation on the chromosome, colonies were isolated by restreaking, and plasmid DNA was purified; this DNA was then used to transform naïve cells of the same auxotroph, and the transformed cells were once again spread on both LB and minimal plates. Growth of approximately the same number of colonies on selective plates as on rich plates indicates that the observed phenotype (growth on minimal) transferred with the genotype (the plasmid carrying the novel gene). These results are shown in Figure 2C and Figure S1. To ensure that rescue was due to a de novo protein, and not some natural sequence that might have been picked up inadvertently during plasmid constructions, we isolated the ∼300 base pair fragment encoding the novel protein, recloned it into a new vector, and showed that the new clone also rescued the deletion strain.
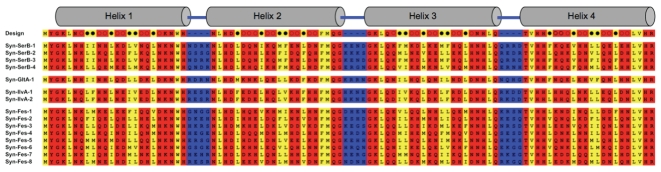
The binary patterned sequences that rescue the four auxotrophs are shown in Figure 3. (Additional sequences are shown in Figure S2.) The novel sequences are designated according to the auxotroph they rescue, followed by a number (e.g. Syn-Fes-1 is a synthetic sequence that rescues Δfes.) For the ΔgltA strain, we isolated one novel protein that enabled cell growth. However, in the other cases, several sequences were isolated, suggesting that deletions of these genes are relatively easy to rescue. As shown by the color-coding in Figure 3, the sequences of the biologically active proteins conform to our binary pattern for the design of 4-helix bundles. (As is often true for sequences in combinatorial libraries, spurious mutations were observed occasionally, but these do not interfere with the overall binary pattern – see Figure S2). NCBI-BLAST searches indicate the sequences of the de novo proteins are unlike those of any known natural proteins [15].
Figure 3. Designed amino acid sequences that enable growth of E. coli auxotrophs on selective media.
Names of the novel (Syn) proteins and the auxotroph rescued are listed at left. The novel sequences follow the designed binary pattern of polar (red) and nonpolar (yellow) residues. The top line shows the template designed to encode four amphiphilic alpha-helices. Combinatorial positions are shown as empty circles (polar residues), filled circles (nonpolar residues), or hyphens (turn residues). Sequences were submitted to the European Nucleotide Archive and assigned accession numbers FR718891 - FR718908.
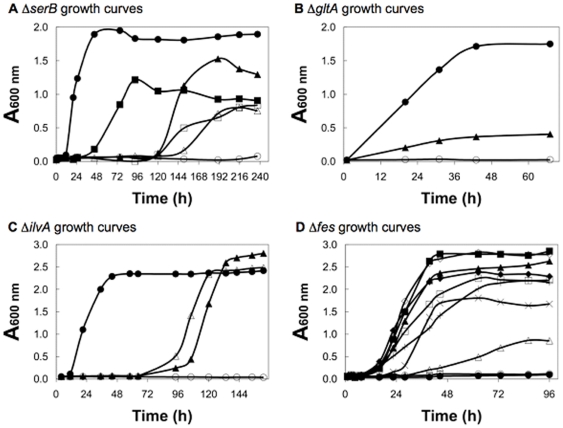
The ability of each de novo protein to sustain cell growth was measured in cultures grown in minimal M9-glucose liquid media. Growth rates were compared for deletion strains expressing a de novo protein versus the same strains expressing either the negative control (LacZ) or the corresponding natural protein on the same pCA24NMAF2 vector. As shown in Figure 4, the novel proteins enable cell growth under conditions where auxotrophs transformed with the control plasmid fail to grow at all. Cells relying on the de novo proteins grow significantly slower than those expressing the natural protein. (Fes is an exception because overexpression of E. coli Fes is toxic [14, Genobase ORF JW0576 - http://ecoli.naist.jp/GB6/info.jsp?id=JW0576].) It is not surprising that the unevolved novel sequences function at substantially lower levels than natural sequences selected by billions of years of evolution. Indeed, the relatively slow rates of cell growth enabled by these first-generation de novo sequences suggest that selections for faster growth might facilitate the evolution of more active proteins. Such experiments will provide an opportunity to test whether de novo progenitor sequences might lead to novel evolutionary trajectories.
Figure 4. Growth of auxotrophic strains of E. coli in selective liquid media.
In each panel, the bottom curve shows that the negative control expressing LacZ (open circles) does not support growth. In contrast, most of the positive controls (black circles) expressing the natural E. coli proteins grow well. (Fes is an exception because overexpression of E. coli Fes is toxic.) The designed proteins enable growth that is well above background. (A) ΔserB: LacZ, open circles; SerB, closed circles; Syn-SerB-1, closed triangles; Syn-SerB-2, open triangles; Syn-SerB-3, closed squares; and Syn-SerB-4, open squares. (B) ΔgltA: LacZ, open circles; GltA, closed circles; and Syn-GltA-1, closed triangles. (C) ΔilvA: LacZ, open circles; IlvA, closed circles; Syn-IlvA-1, closed triangles; and Syn-IlvA-2, open triangles. (D) Δfes: LacZ, open circles; Fes, closed circles; Syn-Fes-1, closed triangles; Syn-Fes-2, open triangles; Syn-Fes-3, closed squares; Syn-Fes-4, open squares; Syn-Fes-5, closed diamonds; Syn-Fes-6, open diamonds; Syn-Fes-7, ×; and Syn-Fes-8, +.
The results shown in Figures 2 and 4 demonstrate that the de novo sequences enable cell growth; however, viability per se does not indicate that the novel proteins provide the same biochemical activities as the deleted natural proteins. Therefore, we devised a series of experiments to probe the functions of the de novo proteins.
Biological functions of the de novo proteins
The natural genes deleted in these four auxotrophs encode a range of functions (9). These are summarized below and depicted graphically in Figure S3.
serB encodes phosphoserine phosphatase, responsible for the final step in serine biosynthesis: phosphoserine → serine + phosphate.
gltA encodes citrate synthase, which catalyzes an early step in glutamate biosynthesis: oxaloacetate + acetyl-coA → citrate → cis-aconitate → isocitrate → alpha-ketoglutarate → glutamate.
ilvA encodes biosynthetic threonine deaminase, which catalyzes the first step in the production of isoleucine from threonine: threonine → 2-ketobutyrate + ammonia → → → isoleucine.
fes encodes enterobactin esterase, which cleaves the iron-bound enterobactin siderophore, thereby enabling cells to acquire iron in iron-limited environments [16].
In principle, the novel proteins could rescue these auxotrophs either by providing the same function as the deleted enzyme, or by one of the three alternative mechanisms described below:
First, we considered the possibility that an auxotroph in a biosynthetic pathway might be rescued by a de novo sequence that produces the end product via a novel bypass pathway. Although it seems unlikely that small, unevolved proteins could catalyze novel biosynthetic pathways, we nonetheless tested this possibility. If the novel sequences that rescue ΔserB, ΔgltA or ΔilvA encode bypass pathways for the synthesis of serine, glutamate, and isoleucine, respectively, then these de novo sequences would also rescue cells deleted for enzymes that function at other steps in the these biosynthetic pathways. (Pathways are shown in Figure S3). This possibility was tested by the following three experiments:
De novo sequences that rescued ΔserB were transformed into ΔserC cells, which are deleted for phosphoserine aminotransferase. This enzyme converts phosphohydroxypyruvate to phosphoserine in the step prior to that catalyzed by the serB encoded enzyme. (Because the serC gene product is also involved in the biosynthesis of pyridoxine [9], the selective media for this experiment was supplemented with pyridoxine.)
The sequence that rescued ΔgltA was transformed into Δicd cells. This strain is also a glutamate auxotroph because it cannot catalyze the conversion of isocitrate to alpha-ketoglutarate, the direct precursor of glutamate.
The sequences that rescued ΔilvA were transformed into ΔilvC and ΔilvD cells, which are deleted for isomeroreductase and dihydroxyacid dehydratase, respectively. Both these enzymes function downstream of ilvA in the biosynthesis of isoleucine. (Because the ilvC and ilvD gene products are also involved in valine biosynthesis [9], the selective media for these experiments was supplemented with valine.)
In all cases, the de novo sequences did not rescue E. coli cells deleted for functions at other steps in the biosynthetic pathways. This demonstrates that the novel proteins do not enable bypass pathways for the synthesis of their respective end products.
Second, we considered the possibility that our de novo sequences might rescue the auxotrophs by altering the expression or activity of an endogenous E. coli protein. To assess this possibility, we relied on an exhaustive screen by Patrick et al., who identified natural genes whose overexpression can rescue the deletion of noncognate genes in E. coli (i.e. ‘multicopy suppressors’) [17]. By screening the complete set of overexpressed E. coli ORFs, they found the following multicopy suppressors of our four strains: ΔilvA can be rescued by overexpression of tdcB [18], or emrD; ΔserB can be rescued by overexpression of gph, hisB, or ytjC; and Δfes can be rescued by overexpression of thiL or setB. ΔgltA was not rescued by overexpression of any E. coli genes. To probe whether our novel sequences rely on these E. coli proteins for rescue, we tested whether our proteins rescue the following double deletion strains: ΔilvAΔtdcB, ΔilvAΔemrD, ΔserBΔgph, ΔserBΔytjC, and ΔfesΔsetB. Transformation with the appropriate novel sequences showed that all five of these double knockouts were rescued by all of the corresponding artificial sequences listed in Figure 3. The ability of our novel sequences to rescue these double deletions shows that the artificial proteins do not function by altering the expression or activity of E. coli proteins known previously to enable rescue.
The double mutant ΔfesΔthiL cannot be constructed because deletion of thiL is lethal. The ΔserBΔhisB strain was constructed, but since ΔhisB is itself an auxotroph, our novel sequences did not rescue this strain (successful rescue would require the unlikely occurrence of a single de novo protein replacing the activities of two conditionally essential genes.) Therefore, dependence on thiL and hisB could not be assessed. In addition, it remains possible that some other E. coli protein, which escaped selection by Patrick et al., could act as a partner in the rescue mediated by our de novo sequences. With the exception of these caveats, our results demonstrate that the known multicopy suppressors are not required for the biological activities of our de novo proteins.
Third, we considered the possibility that the de novo proteins might rescue the deletion strains by a mechanism that does not depend on the specific sequences (Figure 3), but instead involves global alterations in metabolism that are induced by the mere expression of foreign genes. For example, although our proteins were designed to fold into alpha-helical bundles, we considered the possibility that sequences isolated by our selections might be unfolded, and thereby induce a cellular stress response. To assess folding, we purified several proteins and measured their circular dichroism spectra. These spectra (shown in Figure S4) demonstrate the structures are predominantly alpha-helical, and similar to the spectra of designed 4-helix bundles solved previously by NMR [19], [20]. Thus, rescue is not caused by unfolded sequences inducing a stress response.
We also note that if a generic stress response were responsible for rescue, one would expect all the Syn proteins to rescue all the deletions. This is not the case. Syn-Fes does not rescue ΔgltA, Syn-SerB does not rescue ΔilvA, and so on. Thus, specific de novo sequences mediate the rescue of specific chromosomal deletions.
To demonstrate explicitly that rescue depends on a specific sequence, rather than a generic cellular response to the expression of foreign genes, we mutated one of the de novo proteins. The design of this mutant was based on an analysis of the common features among the de novo sequences that rescued ΔilvA. Alignment of the Syn-IlvA sequences revealed two conserved polar residues: Glu36 and Lys42. We constructed the Lys42»Ala mutation in Syn-IlvA-1, and found that although this protein was expressed at the same level as the parental sequence, it fails to rescue ΔilvA. (These results are shown in Figure S5) Thus, rescue is not simply due to expression of an exogenous gene; it is mediated by sequence-specific features of Syn-IlvA-1.
Fes differs from the other deletions because it is not involved in a biosynthetic pathway. Fes functions in iron acquisition. E. coli secretes the enterobactin siderophore (MW, 670 Da) into the media, where it binds iron, and is transported back into the cell. Because the affinity of enterobactin for iron is extremely high (KD≈10−35 M), release of the metal requires degradation of the siderophore. This is catalyzed by the Fes protein, enterobactin esterase [16]. The impact of the de novo syn-fes sequences on iron acquisition is dramatic: Elemental analysis shows that cells expressing the Syn-Fes proteins accumulate 6- to 10-fold more iron than control cells (Figure S6). In principle, the de novo proteins could rescue Δfes either by functioning as an esterase or by some alternative mechanism. For example, the de novo proteins could enable iron acquisition by being transported into the media, binding iron with high affinity, being re-transported back into the cell, and then releasing the tightly bound iron intracellularly. Although possible, this alternative mechanism seems unlikely for novel proteins that occur so frequently in a semi-random library.
Irrespective of whether the de novo proteins rescue auxotrophs by providing the same function as the deleted enzyme or by some alternative mechanism, we expected the artificial sequences – which were neither selected by evolution (in vivo or in vitro), nor explicitly designed for enzymatic activity – to have far lower levels of activity than naturally evolved sequences. To estimate this level of activity in vivo, we compared the growth rates of deletion strains expressing a high level (400X) of Syn-IlvA-1 to the same strain expressing a low level (1X) of the natural protein, encoded by IlvA. These experiments showed that even when the de novo protein is expressed at ∼400-fold higher levels than the natural protein, cells grow much more slowly. (Growth rates are shown in Figure S7.) As expected, the de novo protein exhibits a very low level of biological activity.
Activity levels that are barely sufficient to sustain slow cell growth may not be detectable in vitro. Indeed, others have reported that although overexpression of one E. coli protein can sometimes rescue the deletion of another protein in vivo, these ‘moonlighting’ activities can be so low that they cannot be detected in vitro [21], [22]. Despite these concerns, we attempted to assay biochemical activities in vitro using both cell lysates and purified proteins. Lysates are easy to prepare and are more likely to contain molecular partners (proteins, cofactors, or metals) present in vivo. The disadvantage of lysates, however, is that background activities may obscure the low-level activities of the de novo proteins. We assayed for phosphoserine phosphatase, enterobactin esterase, threonine deaminase, and citrate synthase activities in lysates from cells expressing the respective de novo proteins. In several cases, activity was observed, however, lysates from control cells also showed low levels of activity. This was not surprising, particularly for the Fes and SerB activities, since previous work in our laboratory showed that E. coli lysates contain nonspecific esterases and phosphatases [13].
We also purified several of the de novo proteins. (To avoid contamination by the natural enzyme, purifications were from strains deleted for the natural gene.) We tested these purified proteins for the enzymatic activities deleted in the respective auxotrophs, but were unable to detect activity that was reproducibly above the controls. There are several reasons why such experiments might not demonstrate activity: (i) As noted above, the novel proteins have extremely low levels of activity. (ii) The de novo proteins may require cofactors. We do not know which cofactors might be required, as the novel proteins may use different cofactors than the natural enzymes. (iii) The novel proteins may function by partnering with an E. coli protein. For example, the specificity of an endogenous hydrolase might be altered by binding one of our helical bundles. If this were the case, activity would not be observed in preparations containing only the purified de novo protein. (iv) The Syn protein may function by a different mechanism than the deleted protein, and novel activities would not be detected in experiments designed to assay the natural enzyme.
While we do not yet know the precise mechanisms by which the novel proteins rescue these four deletions in E. coli, we have ruled out several alternative mechanisms including (i) bypass pathways, (ii) activation of known endogenous suppressors, and (iii) induction of a generic stress response (see above). The slow growth rates of cells relying on the novel proteins for survival indicate that regardless of the actual mechanism, the de novo proteins have very low levels of activity — as expected for sequences that were neither designed nor evolved for function. Irrespective of whether the de novo proteins catalyze the same biochemical reaction as the deleted natural protein, or function by some alternative mechanism (and whether they act directly or through interactions with endogenous proteins), elucidating the molecular basis of auxotroph rescue will enhance our understanding of the minimal functions necessary for cell growth.
Replacement of 0.1% of the E. coli genome
We showed above that artificial genes enable cell viability. Next, to assess whether portions of a genome can be replaced by genetic information devised in the laboratory, we probed the ability of several novel sequences to rescue the deletion of several genes simultaneously. A quadruple knockout strain – Δfes ΔgltA ΔilvA ΔserB – was constructed and designated QUAD. This strain has severe growth defects: While the parent strain forms colonies after overnight incubation on either nutrient-rich or nutrient-poor media, QUAD requires almost two days to form colonies on rich media, and as expected, does not form colonies on minimal media. To assess the ability of artificial genes to substitute simultaneously for the deleted functions, we transformed QUAD with derivatives of the pQLinkN vector [23] co-expressing four artificial genes shown above to rescue the four single-gene knockouts. The sequences used for this experiment were syn-fes-2, syn-gltA-1, syn-ilvA-1, and syn-serB-2 (Figure 3). Transformed cells were plated on minimal media. As a control, we transformed QUAD with empty vector; no colonies were observed for the control. However, as shown in Figure 5, transformation with the vector expressing all four artificial genes produced colonies. As discussed above, the individual Syn proteins are considerably less active than the deleted natural proteins. Therefore, it is not surprising that the simultaneous rescue of four deletions by four artificial proteins produced small colonies that required 13 days to appear. Nonetheless, the rescue of QUAD demonstrates that novel sequences bearing no similarity to naturally evolved proteins can compensate for deletion of a portion of the E. coli chromosome.
Figure 5. Rescue of a quadruple knockout E. coli by co-expression of 4 de novo proteins.
(A) Schematic of knockout rescue. Top: Wild-type E. coli transformed with empty vector forms colonies on M9/glucose minimal media. Middle: QUAD E. coli transformed with empty vector does not form colonies on minimal media. Bottom: QUAD transformed with a vector co-expressing four de novo sequences forms colonies on minimal media. (B) Colonies are not observed when QUAD is transformed with empty vector (top panel), but are observed when transformed with a vector expressing four artificial genes (bottom panel). Scale bar = 1 mm.
The E. coli genome contains approximately 4,000 genes [5]. Of these, only 290 are essential for survival under all conditions [24]. An additional 107 are required for growth in the nutrient-poor media provided by M9 salts with glucose [17]. Thus, only ∼400 genes are essential for growth in minimal media. Our results demonstrate that at least 1% of the E. coli genome that is absolutely required for growth under minimal conditions – and 0.1% of the entire E. coli genome – can be replaced by artificial genes designed de novo.
Conclusion
We have demonstrated that sequences designed de novo can provide functions necessary to sustain the growth of living cells. It should be emphasized that these macromolecules were isolated with relatively high frequency from a collection of sequences that were designed to adopt a stable globular fold [7], [8], [19], [20], [25], but were not explicitly designed for function.
Both in terms of linear sequence and 3-dimensional structure, the novel proteins differ substantially from the natural proteins they replace: The binary patterned sequences in Figure 3 show no significant sequence similarity to any known proteins. At the structural level, the 102-residue 4-helix bundles specified by our design are much smaller and simpler than the structures of the four natural proteins deleted in the auxotrophs. (Structures are shown in Figure S8). Thus, although natural proteomes include diverse structures and topologies, our studies show that in some cases, cell growth can be sustained by much simpler structures.
The field of synthetic biology — and the related field of astrobiology — aim to go beyond questions about “what already exists” and attempt to probe “what might be possible?” The results described here suggest that the toolkit for synthetic biology need not be limited to genes and proteins that already exist in nature. We tested 27 deletion strains using a library of 1.5 million sequences designed to fold into 4-helix bundles. From these, we found 18 artificial sequences (Figure 3 and Figure S2) that compensate for the deletion of 4 different genes, and provide biological functions enabling cell growth. Extension of this work to other deletion strains, using larger libraries, and designs that encode a greater diversity of protein structures can be expected to increase substantially the number of life-supporting functions that can be provided by genes designed de novo. The results described here showing that sequences designed in the laboratory can replace portions of the E. coli genome can therefore be viewed as an initial step toward the construction of artificial genomes.
Materials and Methods
Construction of a library of de novo sequences
The design, construction, and characterization of a library of artificial genes encoding binary-patterned de novo proteins are reported elsewhere [11], [12]. The library of gene inserts was digested with NdeI and BsrGI, as was plasmid pCA24NMAF2. Double-digested plasmid backbone was gel purified, and the library insert and vector backbone were ligated. Ligation products were transformed into electrocompetent XL1-blue (Stratagene) or MDS42recA (Scarab Genomics, LLC). Colonies were harvested by scraping from LB plates supplemented with 34 µg ml-1 chloramphenicol (cam), and plasmid DNA was purified from the resulting cell suspension. Aliquots from this library of plasmids were used to transform the auxotrophs. The size of this library—approximately 1.5×106 different sequences—was estimated by plating serial dilutions of the transformations of the initial ligation products on LB-cam [12].
Isolation of de novo sequences that rescue auxotrophs
The collection of plasmids encoding our library of novel sequences, and two negative control plasmids, pCA24N-lacZ and pCA24NMAF2 (empty vector), were used to transform 27 E. coli auxotrophs. These were chosen from Keio strains that grow on LB, but not on M9-glucose. Following electroporation, cells were outgrown at 37°C, harvested by centrifugation, washed and resuspended in chilled M9 salts, and spread on both LB supplemented with 30 µg ml−1 kanamycin (kan), 34 µg ml−1 cam; and on M9-glucose supplemented with 50 µM IPTG, 30 µg ml−1 kan, 34 µg ml−1 cam. After several days at 33°C, plates were scored for number of colonies. In addition, a 1∶100 dilution of library-transformed cells was spread onto LB-cam plates to measure transformation frequency under conditions that were non-selective for auxotroph growth.
Growth assays
Single colonies were inoculated into LB (30 µg ml−1 kan, 170 µg ml−1 cam). Following overnight growth at 37°C, 1 ml of each culture was harvested by centrifugation at 4°C. Cell pellets were washed and resuspended in 1 ml chilled M9 salts, and the resulting cell suspensions were diluted 100-fold into M9-glucose (50 µM IPTG, 30 µg ml−1 kan, 34 µg ml−1 cam). Cultures were grown at 33°C and assayed by measuring absorbance at 600 nm.
Additional methods are described in Supporting Information Text S1.
Supporting Information
Supplementary materials and methods.
(DOC)
Reconfirmation of hits. Colony counts for re-transformation of ΔilvA cells with several different hits. The histogram summarizes 13 experiments of the type shown in figure 2C. The number of colonies on M9-glucose minimal media (dark blue) is similar to the number on LB rich media (light blue).
(TIFF)
De novo sequences with spurious mutations and corresponding ΔilvA growth curves. (A) Novel sequences with spurious mutations. Red indicates polar residues, yellow indicates nonpolar residues, and turns are in blue. Syn-IlvA-3 and Syn-IlvA-4 are 105-residue proteins differing by a single residue at position 28. Both exhibit a 4 amino acid insertion into helix 1, and a 1 amino acid deletion from helix 4. Syn-IlvA-5 is a 106-residue protein, with an 11 amino acid insertion into the second helix and a 7 amino acid deletion from helix 4. These spurious mutations occur at the DNA level and are due either to errors in oligonucleotide synthesis or rare instances of the mis-assembly of library insert building blocks. As long as such spurious mutations do not affect frame or introduce stop codons, the mutated segments survive preselection, and as shown by our results, can contribute to the synthesis of productive sequences. (B) Growth curves for the ΔilvA strain expressing designed artificial proteins with spurious mutations. The bottom curve shows that the negative control expressing LacZ (open circles) does not enable growth. In contrast, the positive control expressing IlvA (closed circles) grows well. Like the novel sequences that adhere precisely to the design, the designed proteins with spurious mutations enable growth that is well above background. Growth curves correspond to the sequences in A as follows: Syn-IlvA-3, closed squares; Syn-IlvA-4, open squares; and Syn-IlvA-5, closed diamonds.
(TIFF)
Reactions and pathways of the deleted proteins. (A) serB encodes phosphoserine phosphatase, responsible for the final step in serine biosynthesis. (B) ilvA encodes biosynthetic threonine deaminase, which catalyzes the first step in the production of isoleucine from threonine. (C) fes encodes enterobactin esterase, which cleaves iron-bound enterobactin. (D) gltA encodes citrate synthase, which catalyzes an early step in glutamate biosynthesis.
(TIFF)
Circular dichroism spectra. CD spectra demonstrate that purified de novo proteins fold into alpha-helical structures. Cultures of E. coli were grown to an OD600 of approximately 0.6 and induced with IPTG. Protein was extracted from the cells by repeated cycles of freezing and thawing. Purification of the Syn proteins to 95% (coommaise-stained SDS-PAGE) was achieved by cation exchange chromatography. Proteins were dialyzed into phosphate buffer for CD analysis.
(TIFF)
Mutation of Lys42 to Ala destroys the ability of a synthetic protein to rescue ΔilvA. (A) Sequence alignment of the six synthetic proteins that rescue ΔilvA. Two charged residues are conserved, E and K (red arrows). Green lines mark regions that are conserved in the designed template. (B) Transformation of ΔilvA cells with Syn-IlvA-1 and Syn-IlvA-1-K42A. Cells expressing Syn-IlvA-1 grow on M9-glucose, but cells expressing Syn-IlvA-1-K42A do not. (C) SDS-PAGE of protein levels after induction with IPTG. Levels of Syn-IlvA-1-K42A are approximately equivalent to those of Syn-IlvA-1. Red arrow marks the band corresponding to the synthetic protein.
(TIFF)
Assay for iron using inductively coupled plasma (ICP) spectrometry. The Δfes strain was transformed with a plasmid directing the high level expression of either LacZ or one of the artificial sequences that rescued the fes deletion. Cells were grown in LB, harvested, and tested for iron accumulation. The artificial proteins enable the accumulation of 6-10 fold more iron than the control.
(TIFF)
De novo proteins are considerably less active than the natural proteins they replace. (A) Growth on minimal media of ΔilvA cells expressing different sequences. Cells expressing the natural IlvA protein grow well either in the presence (Blue) or absence (Red) of IPTG. In contrast, cells expressing Syn-IlvA-1 grow significantly more slowly, even in the presence of IPTG (Green). [These cells do not grow in the absence of IPTG.] Control cells expressing LacZ fail to grow even in the presence of IPTG (Purple). All proteins were expressed off the pCA24N plasmid. (All curves except the red curve represent experiments done in the presence of 0.5 mM IPTG.) (B) Beta-galactosidase assays demonstrate that IPTG induces 402 (+/−7) fold increase in protein expression. LacZ was expressed from the pCA24N plasmid, either in the presence or the absence of IPTG, and levels of beta-galactosidase (Y-axis) were assayed according to standard procedures. The data summarized in this figure show that the Syn-IlvA protein is substantially less active than natural IlvA. The beta-galactosidase assays in panel B demonstrate that IPTG causes a 402 (+/−7) fold increase in protein expression. The results shown in panel A show that for the natural IlvA protein, this change in expression level has no effect on the rate of cell growth: Even low level expression in the absence of IPTG is sufficient for the natural protein to sustain robust growth. In contrast, cells that depend on Syn-IlvA grow slowly even when expression is induced 400-fold. Since cells relying on 400x levels of Syn-IlvA require significantly more time to grow than cells expressing 1x levels of the natural protein, the artificial protein is <1/400 as active as the natural protein. These experiments demonstrate that the artificial proteins are substantially less active in their ability to sustain cell growth. However, they do not indicate that the artificial proteins function by the same mechanism as the natural protein. Indeed, the reduced ability of the artificial proteins to sustain cell growth may be due to their functioning by a less effective mechanism.
(TIFF)
Structures of the 4 natural proteins compared with protein S-824, a de novo 4-helix bundle from a binary patterned library. Coordinates are from the PDB. (A) M. jannaschii SerB (1f5s). (B) E. coli GltA (1k3p). (C) The asymmetric unit of E. coli IlvA (1tdj). (D) S. flexneri Fes (3c87). (E) De novo protein S-824 (1p68).
(TIFF)
Acknowledgments
We thank Wayne Patrick and Ichiro Matsumura for providing materials and advice at the outset of the project; Natividad Ruiz and Tom Silhavy for providing access to the Keio collection, and for guidance about creating the multiple knockouts; Joshua Lees and Fred Hughson for advice on the pQLinkN vector; Josh Rabinowitz and John Groves for advice; and Izhack Cherny for advice and encouragement.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by NSF grant MCB-0817651 (URL: http://www.nsf.gov/div/index.jsp?div=MCB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pereto J, Catala J. The renaissance of synthetic biology. Biol Theor. 2007;2:128–130. [Google Scholar]
- 2.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 3.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52 –56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 4.Beasley JR, Hecht MH. Protein design: The choice of de novo sequences. J Biol Chem. 1997;272:2031–2034. doi: 10.1074/jbc.272.4.2031. [DOI] [PubMed] [Google Scholar]
- 5.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 7.Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 8.Hecht MH, Das A, Go A, Bradley LH, Wei Y. De novo proteins from designed combinatorial libraries. Protein Sci. 2004;13:1711–1723. doi: 10.1110/ps.04690804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson DL, Cox MM. New York, W.H. Worth Publishers; 2008. Lehninger, Principles of Biochemistry 5th ed. [Google Scholar]
- 10.Xiong H, Buckwalter BL, Shieh HM, Hecht MH. Periodicity of polar and nonpolar amino acids is the major determinant of secondary structure in self-assembling oligomeric peptides. Proc Natl Acad Sci USA. 1995;92:6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradley LH, Kleiner RE, Wang AF, Hecht MH, Wood DW. An intein-based genetic selection allows the construction of a high-quality library of binary patterned de novo protein sequences. Protein Eng Des Sel. 2005;18:201–207. doi: 10.1093/protein/gzi020. [DOI] [PubMed] [Google Scholar]
- 12.Bradley LH, Wang AF, Platt J, Fisher MA, Korolev M, Hecht MH. A designed artificial proteome for high throughput screens and selections. In preparation.
- 13.Patel S, Bradley LH, Jinadasa S, Hecht MH. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Protein Sci. 2009;18:1388–1400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, et al. Complete set of ORF clones of Escherichia coli ASKA library (A complete Set of E. coli K-12 ORF Archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Raymond KN, Dertz EA, Kim SS. Enterobactin: An archetype for microbial iron transport. Proc Natl Acad Sci USA. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patrick WM, Quandt E, Swartzlander D, Matsumura I. Multicopy suppression underpins metabolic evolvability. Mol Biol Evol. 2007;24:2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umbarger HE, Brown B. Threonine deaminase in Escherichia coli: Evidence for two L- threonine deaminases. J Bacteriol. 1957;73:105–112. doi: 10.1128/jb.73.1.105-112.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Y, Kim S, Fela D, Baum J, Hecht MH. Solution structure of a de novo protein from a designed combinatorial library. Proc Natl Acad Sci USA. 2003;100:13270–13273. doi: 10.1073/pnas.1835644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go A, Kim S, Baum J, Hecht MH. Structure and dynamics of de novo proteins from a designed superfamily of 4-helix bundles. Protein Sci. 2008;17:821–832. doi: 10.1110/ps.073377908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morett E, Saab-Rincón G, Olvera L, Olvera M, Flores H, et al. Sensitive Genome-Wide Screen for Low Secondary Enzymatic Activities: The YjbQ Family Shows Thiamin Phosphate Synthase Activity. J Mol Biol. 2008;376:839–853. doi: 10.1016/j.jmb.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 22.Patrick WM, Matsumura I. A study in molecular contingency: Glutamine phosphoribosylpyrophosphate amidotransferase is a promiscuous and evolvable phosphoribosylanthranilate isomerase. J Mol Biol 377, 2008;323–336 doi: 10.1016/j.jmb.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheich C, Kummel D, Soumailakakis D, Heinemann U, Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudd KE. EcoGene: A genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Y, Liu T, Sazinsky SL, Moffet DA, Pelczer I, et al. Stably folded de novo proteins from a designed combinatorial library. Protein Sci. 2003;12:92–112. doi: 10.1110/ps.0228003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods.
(DOC)
Reconfirmation of hits. Colony counts for re-transformation of ΔilvA cells with several different hits. The histogram summarizes 13 experiments of the type shown in figure 2C. The number of colonies on M9-glucose minimal media (dark blue) is similar to the number on LB rich media (light blue).
(TIFF)
De novo sequences with spurious mutations and corresponding ΔilvA growth curves. (A) Novel sequences with spurious mutations. Red indicates polar residues, yellow indicates nonpolar residues, and turns are in blue. Syn-IlvA-3 and Syn-IlvA-4 are 105-residue proteins differing by a single residue at position 28. Both exhibit a 4 amino acid insertion into helix 1, and a 1 amino acid deletion from helix 4. Syn-IlvA-5 is a 106-residue protein, with an 11 amino acid insertion into the second helix and a 7 amino acid deletion from helix 4. These spurious mutations occur at the DNA level and are due either to errors in oligonucleotide synthesis or rare instances of the mis-assembly of library insert building blocks. As long as such spurious mutations do not affect frame or introduce stop codons, the mutated segments survive preselection, and as shown by our results, can contribute to the synthesis of productive sequences. (B) Growth curves for the ΔilvA strain expressing designed artificial proteins with spurious mutations. The bottom curve shows that the negative control expressing LacZ (open circles) does not enable growth. In contrast, the positive control expressing IlvA (closed circles) grows well. Like the novel sequences that adhere precisely to the design, the designed proteins with spurious mutations enable growth that is well above background. Growth curves correspond to the sequences in A as follows: Syn-IlvA-3, closed squares; Syn-IlvA-4, open squares; and Syn-IlvA-5, closed diamonds.
(TIFF)
Reactions and pathways of the deleted proteins. (A) serB encodes phosphoserine phosphatase, responsible for the final step in serine biosynthesis. (B) ilvA encodes biosynthetic threonine deaminase, which catalyzes the first step in the production of isoleucine from threonine. (C) fes encodes enterobactin esterase, which cleaves iron-bound enterobactin. (D) gltA encodes citrate synthase, which catalyzes an early step in glutamate biosynthesis.
(TIFF)
Circular dichroism spectra. CD spectra demonstrate that purified de novo proteins fold into alpha-helical structures. Cultures of E. coli were grown to an OD600 of approximately 0.6 and induced with IPTG. Protein was extracted from the cells by repeated cycles of freezing and thawing. Purification of the Syn proteins to 95% (coommaise-stained SDS-PAGE) was achieved by cation exchange chromatography. Proteins were dialyzed into phosphate buffer for CD analysis.
(TIFF)
Mutation of Lys42 to Ala destroys the ability of a synthetic protein to rescue ΔilvA. (A) Sequence alignment of the six synthetic proteins that rescue ΔilvA. Two charged residues are conserved, E and K (red arrows). Green lines mark regions that are conserved in the designed template. (B) Transformation of ΔilvA cells with Syn-IlvA-1 and Syn-IlvA-1-K42A. Cells expressing Syn-IlvA-1 grow on M9-glucose, but cells expressing Syn-IlvA-1-K42A do not. (C) SDS-PAGE of protein levels after induction with IPTG. Levels of Syn-IlvA-1-K42A are approximately equivalent to those of Syn-IlvA-1. Red arrow marks the band corresponding to the synthetic protein.
(TIFF)
Assay for iron using inductively coupled plasma (ICP) spectrometry. The Δfes strain was transformed with a plasmid directing the high level expression of either LacZ or one of the artificial sequences that rescued the fes deletion. Cells were grown in LB, harvested, and tested for iron accumulation. The artificial proteins enable the accumulation of 6-10 fold more iron than the control.
(TIFF)
De novo proteins are considerably less active than the natural proteins they replace. (A) Growth on minimal media of ΔilvA cells expressing different sequences. Cells expressing the natural IlvA protein grow well either in the presence (Blue) or absence (Red) of IPTG. In contrast, cells expressing Syn-IlvA-1 grow significantly more slowly, even in the presence of IPTG (Green). [These cells do not grow in the absence of IPTG.] Control cells expressing LacZ fail to grow even in the presence of IPTG (Purple). All proteins were expressed off the pCA24N plasmid. (All curves except the red curve represent experiments done in the presence of 0.5 mM IPTG.) (B) Beta-galactosidase assays demonstrate that IPTG induces 402 (+/−7) fold increase in protein expression. LacZ was expressed from the pCA24N plasmid, either in the presence or the absence of IPTG, and levels of beta-galactosidase (Y-axis) were assayed according to standard procedures. The data summarized in this figure show that the Syn-IlvA protein is substantially less active than natural IlvA. The beta-galactosidase assays in panel B demonstrate that IPTG causes a 402 (+/−7) fold increase in protein expression. The results shown in panel A show that for the natural IlvA protein, this change in expression level has no effect on the rate of cell growth: Even low level expression in the absence of IPTG is sufficient for the natural protein to sustain robust growth. In contrast, cells that depend on Syn-IlvA grow slowly even when expression is induced 400-fold. Since cells relying on 400x levels of Syn-IlvA require significantly more time to grow than cells expressing 1x levels of the natural protein, the artificial protein is <1/400 as active as the natural protein. These experiments demonstrate that the artificial proteins are substantially less active in their ability to sustain cell growth. However, they do not indicate that the artificial proteins function by the same mechanism as the natural protein. Indeed, the reduced ability of the artificial proteins to sustain cell growth may be due to their functioning by a less effective mechanism.
(TIFF)
Structures of the 4 natural proteins compared with protein S-824, a de novo 4-helix bundle from a binary patterned library. Coordinates are from the PDB. (A) M. jannaschii SerB (1f5s). (B) E. coli GltA (1k3p). (C) The asymmetric unit of E. coli IlvA (1tdj). (D) S. flexneri Fes (3c87). (E) De novo protein S-824 (1p68).
(TIFF)