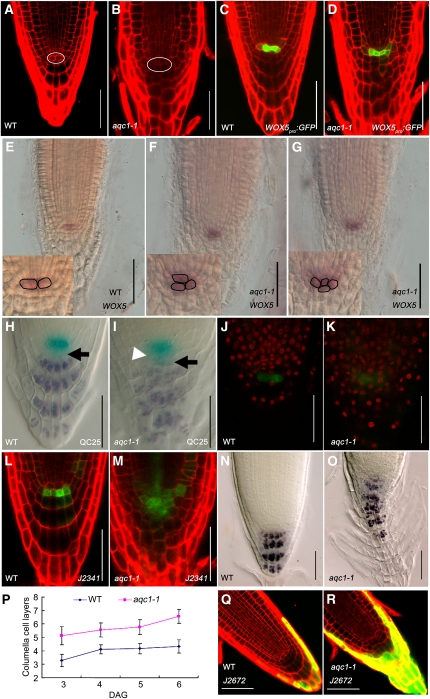
Figure 2.
The aqc1-1 Mutant Shows Extra QC Cells and Defective Root Stem Cell Niche Maintenance.
(A) and (B) Cellular organization of wild-type (WT) (A) and aqc1-1 (B) root tips at 4 DAG using propidium iodide staining. The white oval shows the QC cells.
(C) and (D) Expression pattern of WOX5pro:GFP in the wild type (C) and aqc1-1 (D) at 4 DAG.
(E) to (G) Whole-mount in situ hybridization with a WOX5 probe in wild-type (E) and aqc1-1 seedlings ([F] and [G], different individuals showing one or two extra QC cells) at 5 DAG. Insets show aqc1-1 roots contain supernumerary QC cells compared with the wild type.
(H) and (I) Double staining of the QC25 GUS marker (light blue) and starch granules (dark brown) in the wild type (H) and aqc1-1 (I) at 4 DAG. Black arrows, putative columella stem cell layer; white arrowhead, extra QCs containing starch granules.
(J) and (K) EdU incorporation assays in wild-type (J) and aqc1-1 (K) root meristems. Red fluorescent EdU positive nuclei in the QC cells of aqc1-1 indicate that cell division was activated in these cells in contrast with the wild type.
(L) and (M) Expression pattern of marker J2341 in the wild type (L) and aqc1-1 (M) at 4 DAG.
(N) and (O) Lugol staining of the wild type (N) and aqc1-1 (O) at 5 DAG.
(P) Number of columella cell layers of the wild type and aqc1-1 from 3 to 6 DAG. Data shown are average and sd (n = 18).
(Q) and (R) Expression pattern of marker J2672 in the wild type (Q) and aqc1-1 (R) at 4 DAG.
Bars = 50 μm.