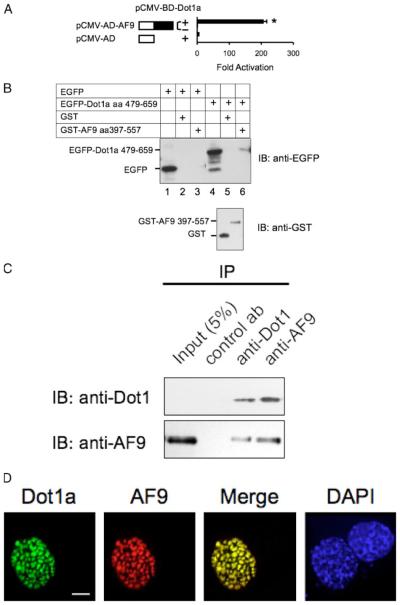
FIGURE 2. Dot1a and AF9 interact in vitro and in vivo.
A, mammalian two-hybrid assay confirming the in vivo interaction of Dot1a and AF9. AF9 was expressed as GAL4 activation domain fusion, and tested to activate a GAL4-dependent luciferase reporter in the presence (+) or absence (−)of GAL4 binding domain fused to Dot1a in mIMCD3 cells. The bars represent the average -fold of activation from three independent experiments. *, p<0.05 versus other bars. B, GST pulldown assay showing specific domains in Dot1a and AF9 responsible for the interaction. GST and GST-AF9-(397–557) fusion were purified from E. coli and incubated with whole cell lysates of mIMCD3 cells expressing EGFP or EGFP-Dot1a-(479–659), as indicated. Input of the lysates (10%, lanes 1 and 4) and proteins bound to glutathione-Sepharose 4B beads wereexamined by immunoblot (IB) analysis with the anti-EGFP antibody. The inputs of GST and GST-AF9 fusion were analyzed similarly with the anti-GST antibody (lower panel). C, co-immunoprecipitation assay demonstrating that the endogenous Dot1 and AF9 proteins in mIMCD3 cells are present in the same protein complex. Whole cell lysates of mIMCD3 cells were immunoprecipitated (IP) with rabbit antibodiesspecific for Dot1, AF9, or H+,K+ -ATPase α2 subunit (as a negative control antibody) as detailed under “Materials and Methods.” Immunoprecipitated proteins were eluted from the protein A/G-agarose beads and subjected to immunoblot analysis in parallel with the antibodies as indicated. D, deconvolution microscopy revealing nuclear colocalization of Dot1a and AF9. mIMCD3 cells were cotransfected with constructs encoding EGFP-Dot1a and red fluorescent protein-tagged AF9 (RFP-AF9) and examined by deconvolution microscopy as detailed under “Materials and Methods.” DAPI, 4′,6-diamidino-2-phenylindole. Scalebar,3 μm.