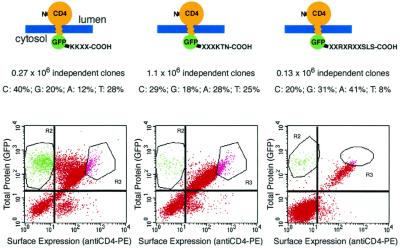
Figure 2.
Libraries of combinatorial variants of di-lysine and arginine-based ER retention/retrieval signals. Schematic depiction of the three libraries analyzed, the randomized positions are indicated in the C-terminal tail. The frequencies of the individual bases were determined by sequencing six to eight randomly picked clones from the unselected libraries. Populations of 3T3 cells infected with each library were sorted on a FACS Vantage SE cell sorter. Gates R2 and R3 were chosen to separate cells according to the two trafficking phenotypes of the reporter protein (retained or expressed on the surface). The plots shown are gated on viable cells as determined by propidium iodide staining (gate R1, not shown).