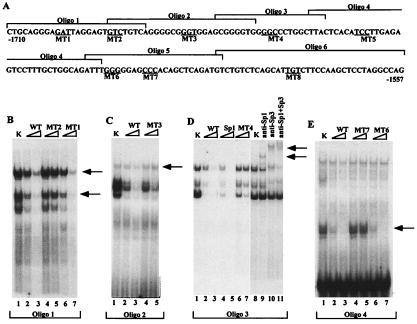
Figure 4.
EMSAs of nuclear extracts showing binding of transcription factors to the HS III region. (A) The upper-strand sequences of the oligonucleotides (1–6) are shown. The mutated sequences are underlined. MT1 (GAT > CGC), MT2 (GTC > AAA), MT3 (GGT > AAA), MT4 (GGC > AAA), MT5 (TCC > AAA), MT6 (GGG > AAA), MT7 (CCC > AAA), and MT8 (TGT > AAA). MT5 and -8 were used in experiments described in Fig. 5. (B) EMSAs were performed with radiolabeled oligonucleotide 1 and nuclear extracts from keratinocytes (K). Competition experiments with different oligonucleotides were performed by using radiolabeled oligonucleotide 1 (WT) and a 20- or 100-fold excess of unlabeled double-stranded oligonucleotides (WT, MT1, and MT2) as indicated over each lane. (C) EMSAs were performed as in B. Arrow indicates the specific complex. (D) EMSAs were performed by incubating oligonucleotide 3 with keratinocyte nuclear extracts. Competitor used in each EMSA is indicated over each lane. Arrows indicate supershifts, reflective of complexes between Sp1 and/or Sp3, DNA, and antibody. Consensus Sp1 oligonucleotides were obtained from Promega. (E) EMSAs were performed with oligonucleotide 5. Specific complex is shown with an arrow.