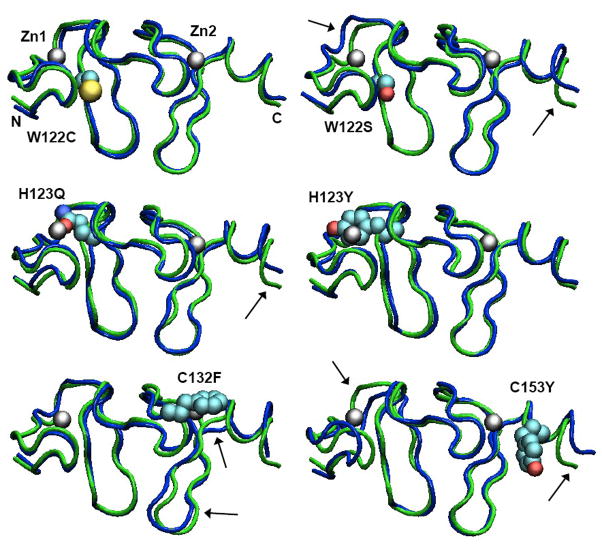
Figure 4.
Structures of the second LIM domain from MD simulations of wild-type FHL1 and the W122C, W122S, H123Y, H123Q, C132F and C153Y mutants. Each structure shown is the mean of 625 structures saved at 20-ps intervals during the trajectory. A tube representation of the wild-type protein is shown in green in each panel, with the indicated mutant structure superimposed in blue. The mutant sidechain and zinc atoms are shown in space-filling (van der Waals) representations colored by atom type. The zinc atoms and the N- and C-termini of the model are labeled in the W122C structure. Over all, the mutant proteins remain folded in essentially the native conformation, with their Zn sites almost fully intact. A few subtle changes are indicated by arrows. The figures were made with VMD 1.8.7 [24].