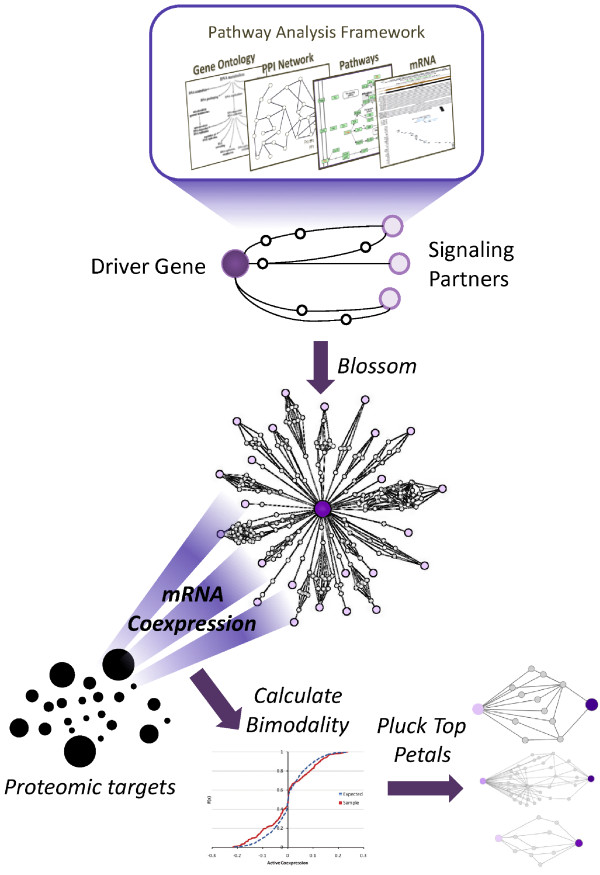
Figure 1.
Workflow for PETALS: Proteomic Evaluation and Topological Analysis of a Mutated Locus' Signaling. We begin with a cancer driver gene of interest (e.g. Apc) and a set of putative signaling partners. The pathways between these two sets are predicted based on protein-protein interactions, coupled with mRNA coexpression and GO annotations. The prediction of pathways to the various signaling partners allows individual subnetworks to blossom into a flower with many petals. The biological relevance of each petal is assessed against proteomic evidence (i.e. 2D-DIGE), using the bimodality of mRNA coexpression to quantify this association. This results in a ranking of petals, which can then be plucked for further experimental evaluation.