Abstract
Cryptosporidium muris, predominantly a rodent species of Cryptosporidium, is not normally considered a human pathogen. Recently, isolated human infections have been reported from Indonesia, Thailand, France, and Kenya. We report the first case of C. muris in a human in the Western Hemisphere. This species may be an emerging zoonotic pathogen capable of infecting humans.
Keywords: Cryptosporidium muris, Perú, zoonotic diseases
Cryptosporidiosis can be a debilitating diarrheal disease. While infections are normally acute and self-limiting in immunocompetent persons, cryptosporidiosis can be life threatening in those with compromised immune systems. In humans, cryptosporidiosis is caused predominantly by Cryptosporidium parvum or C. hominis (the latter was previously known as the C. parvum human genotype), and major outbreaks of the disease have been clearly associated with contaminated drinking water (1).
Recently, another species of Cryptosporidium, C. muris, has been suggested to be of concern to human health. C. muris is a parasite first identified in the gastric glands of mice (2). Experimental transmission studies have shown that the parasite readily infects multiple nonrodent hosts including dogs, rabbits, lambs, and cats (3). C. muris–like organisms have also been reported as opportunistic infectious agents in immunocompromised nonhuman primates (4). In the past 2 years, five cases of infections with C. muris or C. muris–like parasites have been reported from HIV-positive and healthy persons in Kenya (5), France (6), Thailand (7), and Indonesia (8). In this paper, we report on the first documented case of C. muris in a human in the Western Hemisphere. The parasite was recovered during the summer of 2002 in stools of an HIV-positive Peruvian woman with severe diarrhea. This finding was confirmed by light microscopy, polymerase chain reaction (PCR)–restriction fragment length polymorphism (RFLP), and DNA sequencing.
The Study
In 2002, we conducted a year-long collaborative study on the epidemiology of Cyclospora cayetanensis infections in Perú. As part of that study, we collected approximately 100 stool samples in 2.5% potassium dichromate solution from persons in Lima and Iquitos with Cyclospora infection. Fecal samples were initially identified as Cyclospora-positive in Lima, and then transported to the United States for additional confirmation using wet mount and Nomarski interference contrast microscopy.
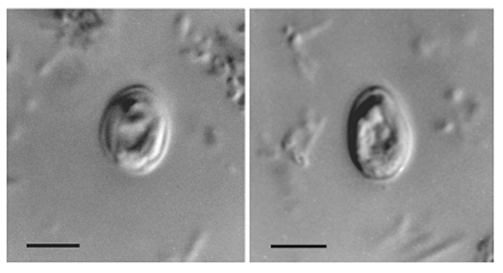
Two stool samples, which were taken on two sequential days from an HIV-positive woman who was 31 years of age, contained oocysts that appeared, based on morphology, to be Cryptosporidium muris. Low numbers of Cyclospora cayetanensis and Blastocystis hominis oocysts were also identified in the stool samples. The Cryptosporidium muris infection was initially identified by using wet mount microscopy with oocysts (n=25) averaging 6.1 (± 0.3) x 8.4 (± 0.3) μM (range 5.6–6.4 x 8.0–9.0) and a shape index (length/width) 1.38 (1.25–1.61) (Figure 1). Numbers of oocysts were determined semiquantitatively in each sample by hemacytometer, with an estimated 737,000 and 510,000 oocysts/g recovered from the submitted samples on day 1 and day 2, respectively. The diagnosis of C. muris was later confirmed through DNA analysis.
Figure 1.
Nomarski interference contrast photomicrographs of Cryptosporidium muris from the feces of an HIV-positive human. Scale bars = 5 μm.
HIV was first diagnosed in the patient in November 2000 by using enzyme-linked immunosorbent assay and Western blot (immunoblot). She arrived at the hospital clinic in June 2002 with fever and reported that she had been experiencing diarrhea for >3 months. The patient reported that she had lost approximately 25 lbs. in the past 7 months, consistent with HIV-wasting syndrome. Her chest x-ray was abnormal, but four direct sputum examinations for acid-fast bacteria using Ziehl-Neelsen staining were negative, as were efforts at culturing Mycobacterium tuberculosis.
Other laboratory values for this patient at the time of stool sample collection were as follows: CD4 cell count 66/μL; hematocrit 36%; leukocytes 4,100/μL with 4% bands, 55% neutrophils, 27% lymphocytes, and 0% eosinophils; urine examination normal; creatine 0.8 mg/dL; urea 21 mg/dL; glucose 105 mg/dL; serum glutamic oxalacetic transaminase 30 IU/L; serum glutamic pyruvic transaminase 46 IU/L; and bilirubin 0.9 mg/dL.
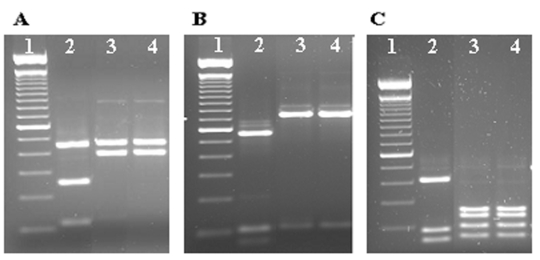
The diagnosis of Cryptosporidium in the patient’s samples was confirmed by a small subunit rRNA-based nested PCR, which amplified a portion of the rRNA gene (830 bp). Cryptosporidium spp. was determined by the banding patterns of restriction digestions of PCR products with SspI, VspI, and DdeI (9). Diagnosis was confirmed by DNA sequencing of three independent PCR products from each sample in both directions on an ABI PRISM 3100 (Applied Biosystems, Foster City, CA) instrument. Figure 2 shows the RFLP analysis of three PCR products from each sample with restriction enzymes SspI and VspI; these results suggest that these PCR products belonged to either C. muris or Cryptosporidium andersoni (10). Further RFLP analysis with DdeI showed banding patterns identical to C. muris (9; Figure 2). All DNA sequences obtained from the six PCR products were identical to those previously reported by Xiao et al. (10,11) from C. muris from a Bactrian camel, a rock hyrax, and mice (GenBank accession nos. AF093997 and AF093498) and another isolate recently found in an HIV patient in Kenya (5).
Figure 2.
Identification of Cryptosporidium muris from two stool samples from a Peruvian patient using restriction fragment length polymorphism analysis of polymerase chain reaction products with SspI (A), VspI (B) and DdeI (C). Lane 1, 100-bp molecular markers; lane 2, C. hominis control; lanes 3 and 4, C. muris from the patient.
After the diagnosis of intestinal parasite infection, the patient was treated with TMP-SMX (trimethoprim 160 mg, sulfamethoxazole 800 mg) Forte twice a day for 1 week and then TMP-SMX once a day for Pneumocystis carinii pneumonia prophylaxis. The patient was also placed on AZT/3TC and nevirapine. The patient recovered with no further evidence of Cyclospora, Blastocystis, or C. muris in stool samples taken 2 months posttreatment. She became afebrile and had gained 5 kg as of 2 months’ posttreatment. Molecular analysis of a stool sample collected 122 days after the initial diagnosis confirmed that the patient had recovered from the C. muris infection.
Conclusions
This report represents the third confirmed case of C. muris infection in humans. Previously, one case of C. muris infection was identified in an HIV-positive child in Thailand and in an HIV-positive adult in Kenya using microscopy and molecular analysis (5,7). C. muris and C. andersoni–like oocysts were found in two healthy Indonesian girls, but the diagnosis was not confirmed by molecular tools (8). One putative C. muris infection was reported in an immunocompromised patient in France based on sequence analysis of a small fragment of the SSU rRNA (6). However, the sequence presented was more similar to that of C. andersoni (2-bp differences in a 242-bp region) than to C. muris (8-bp differences in the region).
Although determining whether or not the C. muris contributed medical problems in this patient is not possible, detecting C. muris in her stool sample is an unexpected finding. A major difference between C. parvum or C. hominis and C. muris, is that C. parvum and C. hominis normally colonize the intestine, whereas C. muris is a gastric pathogen in cattle. Anderson (12) and Esteban and Anderson (13) reported that another gastric species, C. andersoni, infects only the glands of the cattle stomach (abomasum), where it retards acid production. These researchers postulated that this process may affect protein digestion in the abomasum and account for the fact that milk production in cows that are chronically infected with C. muris appears to be reduced by approximately 13%. Thus, an infection by C. muris may perhaps cause similar protein digestion problems in human infections, particularly in HIV-positive persons.
Even though only a few cases of C. muris infections have been identified so far in humans, gastric cryptosporidiosis occurs much more often than believed, especially in HIV-positive persons. Up to 40% of cryptosporidiosis in HIV-infected persons includes gastric involvement (14). Although most gastric Cryptosporidium infections in HIV-positive persons are likely caused by C. parvum or C. hominis because of immunosuppression, the contribution of C. muris probably has been underestimated. Thus, molecular characterizations of stomach tissues from patients with gastric cryptosporidiosis may help us to understand the pathogenesis of human Cryptosporidium infection.
Our report expands the geographic range of suspect C. muris infections in humans and suggests that this species may be a global emerging zoonotic pathogen. This pathogen may be of particular importance to persons living in regions where rodents live in close proximity to humans and sanitation may be minimal. C. muris may also be more prevalent than currently recognized. The organism is nearly twice as large as C. parvum and closer in size to Cyclospora cayetanensis. Although Cyclospora autofluoresces while Cryptosporidium does not (15), C. muris could still be easily misdiagnosed, since few laboratory workers would be familiar with C. muris or its morphology.
Acknowledgments
We thank the laboratory support personnel at the Instituto de Medicina Tropical Alexander von Humboldt, Universidad Peruana Cayetano Heredia, Lima, Perú, especially Jenny Anchiraico.
This study was supported by Environmental Protection Agency award numbers R-82858602-0 (C.J.P.) and R-82837001-0 to (S.J.U.) and by a research grant (L.X.) from the Opportunistic Infections Working Group at the Centers for Disease Control and Prevention, Atlanta, GA.
Biography
Dr. Palmer is a research professor at the University of Florida. Her primary research interests are infectious and tropical disease with special emphasis on field-based research studies in the Americas.
Footnotes
Suggested citation for this article: Palmer CJ, Xiao L, Terashima A, Guerra H, Gotuzzo E, Saldías G, et al. Cryptosporidium muris, a rodent pathogen, recovered from a human in Perú. Emerg Infect Dis [serial online] 2003 Sept [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol9no9/03-0047.htm
References
- 1.Fayer R, Morgan U, Upton SJ. Epidemiology of Cryptosporidium: transmission, detection and identification. Int J Parasitol. 2000;30:1305–22. 10.1016/S0020-7519(00)00135-1 [DOI] [PubMed] [Google Scholar]
- 2.Tyzzer EE. An extracellular coccidium, Cryptosporidium muris (gen. et sp. nov.), of the gastric glands of the common mouse. Arch Protistenkd. 1910;26:394–418. [PMC free article] [PubMed] [Google Scholar]
- 3.Izeki M, Maekawa T, Moriya K, Uni S, Takada S. Infectivity of Cryptosporidium muris (strain RN 66) in various laboratory animals. Parasitol Res. 1989;75:218–22. 10.1007/BF00931279 [DOI] [PubMed] [Google Scholar]
- 4.Dubey JP, Markovitis JE, Killary KA. Cryptosporidium muris–like infection in stomach of cynomolgus monkeys (Macaca fascicularis). Vet Pathol. 2002;39:363–71. 10.1354/vp.39-3-363 [DOI] [PubMed] [Google Scholar]
- 5.Gatei W, Ashford RW, Beeching NJ, Kamwati SK, Greensill J, Hart CA. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg Infect Dis. 2002;8:204–6. 10.3201/eid0802.010256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guyot K, Follet-Dumoulin A, Lelievre E, Sarfati C, Rabodonirina M, Nevez G, et al. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39:3472–80. 10.1128/JCM.39.10.3472-3480.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiangtip R, Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop Med Int Health. 2002;7:357–64. 10.1046/j.1365-3156.2002.00855.x [DOI] [PubMed] [Google Scholar]
- 8.Katsumata T, Hosea D, Ranuh IG, Uga S, Yanagi T, Kohno S. Short report: possible Cryptosporidium muris infection in humans. Am J Trop Med Hyg. 2000;62:70–2. [DOI] [PubMed] [Google Scholar]
- 9.Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol. 2001;67:1097–101. 10.1128/AEM.67.3.1097-1101.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao L, Escalante L, Yang CF, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao L, Sulaiman IM, Ryan UM, Zhou L, Atwill ER, Tischler ML, et al. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int J Parasitol. 2002;32:1773–85. 10.1016/S0020-7519(02)00197-2 [DOI] [PubMed] [Google Scholar]
- 12.Anderson BC. Cryptosporidiosis in bovine and human health. J Dairy Sci. 1998;81:3036–41. 10.3168/jds.S0022-0302(98)75868-0 [DOI] [PubMed] [Google Scholar]
- 13.Esteban E, Anderson BC. Cryptosporidium muris: prevalence, persistency, and detrimental effect on milk production in a drylot dairy. J Dairy Sci. 1995;78:1068–72. 10.3168/jds.S0022-0302(95)76723-6 [DOI] [PubMed] [Google Scholar]
- 14.Lumadue JA, Manabe YC, Moore RD, Belitsos PC, Sears CL, Clark DP. A clinicopathologic analysis of AIDS-related cryptosporidiosis. AIDS. 1998;12:2459–66. 10.1097/00002030-199818000-00015 [DOI] [PubMed] [Google Scholar]
- 15.Varea M, Clavel A, Doiz O, Castillo FJ, Rubio MC, Gómez-Lus R. Fuchsin fluorescence and autofluorescence in Cryptosporidium, Isospora and Cyclospora oocysts. Int J Parasitol. 1998;28:1881–3. 10.1016/S0020-7519(98)00146-5 [DOI] [PubMed] [Google Scholar]