Abstract
The active center of acetylcholinesterase (AChE), a target site for competitive inhibitors, resides centrosymmetric to the subunit at the base of a deep, narrow gorge lined by aromatic residues. At the gorge entry, a peripheral site encompasses overlapping binding loci for non-competitive inhibitors, which alter substrate access to the gorge. The click-chemistry inhibitor TZ2PA6 links the active center ligand, tacrine, to the peripheral site ligand, propidium, through a biorthogonal reaction of an acetylene and an azide that forms either a syn1 or an anti1 triazole. Compared with wild-type mouse AChE, a Tyr337Ala mutant displays full catalytic activity, albeit with two to three orders of magnitude higher affinities for the TZ2PA6 syn1 and anti1 regioisomers, reflected in low femtomolar Kd values, diffusion-limited association and dissociation half-times greater than one month and one week, respectively. Three structures of each of the co-crystallized syn1 and anti1 complexes of the Tyr337Ala mutant were solved at three distinct times of crystal maturation, consistent with or exceeding the half-lives of the complexes in solution, while crystalline complexes obtained from soaked Tyr337Ala crystals led to picturing “freshly formed” complexes. The structures, at 2.55-2.75Å resolution, reveal a range of unprecedented conformations of the bound regioisomers, not observed in the wild-type AChE complexes, associated with concerted positional rearrangements of side chains in the enzyme gorge. Moreover, time-dependent conformational remodeling of the crystalline complexes appears to correlate with the dissociation half-times of the solution complexes. Hence for the tight-binding TZ2PA6 inhibitors, the initial complexes kinetically driven in solution slowly form more stable complexes governed by thermodynamic equilibrium and observable in mature crystals.
Keywords: Acetylcholinesterase, active site gorge, click chemistry, conformational change, crystal structure, femtomolar dissociation constant, peripheral site, triazole
1. Introduction
Acetylcholinesterase (AChE) rapidly terminates cholinergic neurotransmission by catalyzing the hydrolysis of the neurotransmitter, acetylcholine, at peripheral and central synapses.1 Inhibitors of AChE have been used for over a century in various therapeutic strategies in certain neuromuscular, ophthalmic, and cognitive disorders.2 Competitive, reversible or irreversible inhibitors of AChE bind the active center, that contains the Glu/His/Ser catalytic triad and is located centrosymmetric to the molecule at the bottom of a deep and narrow gorge.3 Non-competitive, reversible inhibitors and activators bind the peripheral anionic site (PAS), an allosteric surface site encompassing overlapping subsites located at the gorge entrance.4 Some other inhibitors with an extended structure and harboring two functional binding moieties bind at both sites and occupy the gorge pathway.5,6 Such ligands typically display higher affinities than those of the individual components and simultaneously inhibit ACh hydrolysis and impair PAS functionality.7
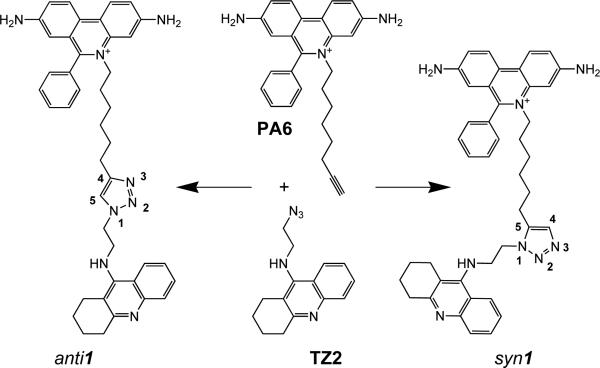
The 1,3-dipolar cycloaddition reaction between azides and terminal alkynes has become a potentially powerful approach for drug discovery.8-11 When reactants are attached to a tetrahydroaminoacridine (tacrine) tricyclic moiety and a phenyl-phenanthridinium moiety (propidium), respectively selective for the active center and PAS of the enzyme, the biorthogonal reaction occurs at a considerable rate if conducted in situ, i.e. inside the active center gorge of AChE (Scheme 1).11,12
Scheme 1. Structures of the anti1 and syn1 TZ2PA6 regioisomers.
The phenyl-phenanthridinium, triazole, and tacrine precursors and moieties are shown from top to bottom. Note the distinctive 1,2,3-triazol-4-yl versus 1,2,3-triazol-5-yl connections for the anti1 and syn1 isomers formed by 1,3-dipolar cycloaddition.11
Moreover, from a series of reagent, building block combinations, the AChE surface preferentially catalyzes the formation of a syn1-triazole regioisomer with defined substitution positions and linker distances. Inhibition measurements revealed this isomer to be a very high affinity, reversible inhibitor of AChE, with association rate constants near the diffusion limit and a sub-picomolar equilibrium dissociation constant (Kd) reflected in the slow dissociation rates for mouse AChE (mAChE) (Table 1). The corresponding anti1-isomer, not formed by the enzyme but generated in vitro, proved to be a respectable though weaker inhibitor with “only” near-picomolar Kd value for mAChE. Triazole formation, midway down the gorge, required the two building blocks be positioned to accommodate the gorge geometry. This approach also permitted us to identify other motifs, phenyltetrahydroisoquinoline, as candidate peripheral site ligands.13
Table 1.
Inhibition or binding parametersa for mAChE and several of its mutants and for other cholinesterases.
| Enzymes and mutants |
anti1 |
syn1 |
||||
|---|---|---|---|---|---|---|
| kon (1010 M-1 min-1) | koff (10-3 min-1) | Kd (fM) | kon (1010 M-1 min-1) | koff (10-3 min-1) | Kd (fM) | |
| mAChEb | 2.5 | 220 | 8900 | 1.7 | 7.1 | 410 |
| mAChE mutants | ||||||
| Tyr124Glnc | 1.9 | 54 | 2800 | 1.2 | 3.9 | 370 |
| Trp286Alab | 0.87 | 1800 | 210000 | 0.94 | 19 | 2000 |
| Tyr337Alac | 1.6 | 0.066 | 4.1 | 1.3 | 0.013 | 1.0 |
| Tyr72Asn/Tyr124Gln/Trp286Alac | 0.067 | 380 | 570000 | 0.056 | 77 | 140000 |
| | ||||||
| AChE, E. electricusb | 1.8 | 250 | 14000 | 1.5 | 1.5 | 99 |
| (conserved Tyr72, Asp74, Tyr124, Trp286, Tyr337) | ||||||
| AChE, T. californicab | 3.2 | 23 | 720 | 1.4 | 1.1 | 77 |
| (Phe substitution for mAChE Tyr337) | ||||||
| AChE, D. melanogasterb | 3.4 | 58 | 1700 | 2.0 | 72 | 3600 |
| (Glu, Tyr, Met substitutions for mAChE Tyr72, Asp74, Tyr124) | ||||||
| Butyrylcholinesterase, mouseb | 0.69 | 3.2 | 460 | 0.36 | 2.6 | 720 |
| (Asn, Arg, Ala substitutions for mAChE Tyr72, Trp286, Tyr337) | ||||||
means of n ≥ 3 individual values with SD ≤ 30% except for Tyr337Ala where SD ≤ 50%.
from ref14.
this study
Comparative crystallographic analysis of the syn1- and anti1-mAChE complexes revealed distinctive orientations of the phenyl-phenanthridinium moieties associated with compensating conformational changes of Trp286 at the enzyme PAS.14 Moreover, the position of the triazole, along with regioisomer preference, suggested that the phenol side chain of AChE Tyr337 protruding into the active center gorge is a critical determinant of specificity. This prompted us to investigate, by site-directed mutagenesis, the role of some of the conserved aromatic side chains that line the active center gorge and establish direct interactions with the bound anti1 and syn1 isomers, as observed in the structures.14 Following mutagenesis of residue Trp286 at the gorge entrance14, we selectively substituted the facing residues Tyr124 and Tyr337 in the constricted region in the gorge and residue Tyr72 in the gorge path, and characterized the mutants through kinetic binding assays. Compared to mAChE, mutant Tyr337Ala was found to display two to three orders of magnitude greater affinities for the TZ2PA6 anti1 and syn1 isomers, leading to Kd values in the low femtomolar range (Table 1).
To explore the structural basis for such further increases in otherwise very high affinities, we solved crystal structures of the Tyr337Ala mutant and of its co-crystallized complexes with the two TZ2PA6 isomers. However, instead of solving a single structure from a lone crystal for each complex as is usually done, from crystals of a same “litter” we collected distinctive data sets at increasing crystal maturation times corresponding to shorter than, equal to and longer than the half-time for complex dissociation in solution (t1/2). Structural analysis of the co-crystallized Tyr337Ala complexes and their comparison with the previously solved mAChE complexes provide evidence for a range of torsional/rotational (anti1) and compressive/extensive (syn1) rearrangements of the bound regioisomers associated with compensatory reorientations of side chains within the enzyme gorge, while the time course of conformational remodeling of the complexes in the crystals appears to correlate with the dissociation half-times in solution. Analysis of crystalline anti1 and syn1 complexes obtained from soaked Tyr337Ala crystals led to picturing “freshly formed” complexes. Finally, analysis of a Tyr337Ala crystal soaked with mixed anti1 and syn1 regioisomers led to a predominant syn1 complex, demonstrating preferential trapping of the slowest dissociating ligand present in a mixture of related compounds. Hence for the tight-binding TZ2PA6 inhibitors, the initial complex formed in solution and observable from a soaked crystal, continues to evolve in maturing co-crystals to achieve lower energy states and better fit.
2. Experimental methods
2.1. Preparation and crystallization of the complexes and data collection
The mAChE mutant Tyr337Ala, generated by site-directed mutagenesis and expressed as a soluble protein in HEK-293 cells,15 was purified by affinity chromatography using decamethonium desorption and dialyzed and prepared as previously described for mAChE16. This mutant displays catalytic parameters for ATCh (kcat, Km) similar to values for mAChE.15 The specific activity of a concentrated, sterile-filtered solution was virtually unaltered after one year storage on ice.
The TZ2PA6 anti1 and syn1 isomers, synthesized and purified as tosylate salts as described,11 were kind gifts from K.B. Sharpless and H.C. Kolb (Dept of Chemistry and Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA). Titration of the isomer stock solutions was carried out spectrophotometrically (ε490 nm = 6000 M-1cm-1).17 The complexes for cocrystallization were prepared using Tyr337Ala at ca. 5 mg/ml and a 2-fold molar excess of each inhibitor (overnight equilibration, 4°C). The unbound ligand was removed and the complexes concentrated to ca. 10 mg/ml using ultrafiltration with controlled rincing. Total inhibition of activity of the complexes was assessed spectrophotometrically18 using enzyme concentrations well above the Kd's.
The Tyr337Ala mutant and its preformed anti1 and syn1 complexes were crystallized at 4°C by vapor diffusion using hanging drops (1 μl) and a protein-to-well solution ratio of 1:1 with PEG-600 25-35% (v/v) in 50-100 mM Hepes, pH 6.0-7.0, or with PEG-550 MME 30% (v/v) in 50 mM Na acetate, pH 7.5, as the reservoir solution. The crystals appeared rapidly, grew to a suitable size within a week and showed limited enlargement thereafter. For each of the co-crystallized complexes, three data sets were collected from crystals that appeared at the same time in the same drop (defined as the same “litter” of crystals) but were flash-cooled at distinct maturation times roughly corresponding to one week, one month and ten months (referred to as “1-wk”, “1-mth” and “10-mth” data sets and structures, respectively). For the co-crystallized syn1 complex, two additional data sets diffracting to ca. 3.0 Å resolution were collected after 18 months (i.e., 15 × t1/2) and 3 years (30 × t1/2) of crystal maturation, leading to similar syn1 conformations as observed in the 10-mth structure (data not shown).
Formation of anti1 and syn1 complexes by soaking Tyr337Ala crystals was carried out at 4°C in 20 μl sitting drops made of the well solution supplemented with 50 μM of anti1, syn1, or both for 24-36 hours. Soaks longer than two days led to ill-diffracting crystals. All crystals were directly flash-cooled in the nitrogen gas stream (100°K). They all belonged to the orthorhombic space group P212121 with unit cell dimensions a = 79.7 Å, b = 111.9 Å, c = 226.5 Å. Oscillation images were integrated with DENZO19 and data scaled and merged with SCALA20 (Table 2).
Table 2.
Data collection and refinement statistics.
| Structure | Tyr337Ala | Co-crystallized complexes |
Soaked complexes |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isomer |
anti1 |
syn1 |
anti1 |
syn1 |
anti1 + syn1 |
|||||
| Maturation time and structure name | 1-wk | 1-mth | 10-mth | 1-wk | 1-mth | 10-mth | na* | na* | na* | |
| Maturation vs t1/2 for dissociation (ca.) | 1 × t1/2 | 4 × t1/2 | 42 × t1/2 | 0.2 × t1/2 | 0.8 × t1/2 | 8 × t1/2 | 0.15 × t1/2 | 0.03 × t1/2 | na* | |
| Data collection a | ||||||||||
| Beamline (ESRF) | ID29 | ID14-EH2 | ID14-EH1 | ID14-EH4 | ID14-EH2 | ID14-EH3 | ID14-EH4 | ID14-EH3 | ID14-EH3 | ID14-EH3 |
| Wavelength (Å) | 0.979 | 0.933 | 0.933 | 0.975 | 0.933 | 0.931 | 0.975 | 0.931 | 0.931 | 0.931 |
| Resolution range (Å) | 20 - 2.65 | 20 - 2.60 | 20 - 2.55 | 20 - 2.65 | 20 - 2.60 | 20 - 2.65 | 20 - 2.75 | 20 - 2.8 | 20 - 2.7 | 25 - 2.7 |
| Total observations | 204 817 | 235 515 | 255 182 | 233 025 | 228 040 | 222 379 | 204 055 | 256 595 | 276 849 | 273 523 |
| Unique reflections | 57 806 | 63 011 | 66 465 | 58 447 | 61 576 | 59 477 | 52 292 | 49 788 | 55 109 | 54 760 |
| Multiplicity | 3.5 (3.6) | 3.7 (3.7) | 3.8 (3.8) | 4.0 (4.0) | 3.7 (3.7) | 3.7 (3.7) | 3.9 (3.9) | 5.2 (5.1) | 5.0 (4.8) | 5.0 (4.7) |
| Completeness (%) | 99.1 (99.1) | 100 (99.4) | 99.5 (99.5) | 99.7 (99.8) | 98.9 (98.6) | 99.0 (99.0) | 99.8 (99.5) | 99.8 (99.8) | 99.8 (99.8) | 99.7 (99.7) |
| I / σ (I) | 12.5 (4.0) | 11.8 (3.3) | 15.8 (4.2) | 15.4 (3.7) | 13.9 (3.6) | 22.4 (3.6) | 15.2 (3.6) | 16.6 (3.6) | 17.1 (3.6) | 17.3 (3.4) |
| Rsymb | 7.7 (41.3) | 8.8 (46.3) | 5.5 (47.5) | 6.5 (46.8) | 5.9 (46.8) | 5.4 (45.0) | 7.2 (48.2) | 8.9 (54.4) | 8.3 (55.0) | 8.4 (56.2) |
| B Wilson plot (A2) | 57.2 | 61.7 | 58.3 | 68.8 | 66.5 | 65.8 | 71.8 | 72.0 | 65.9 | 68.9 |
| Refinement c | ||||||||||
| R-factor / R-free (%) | 19.3 / 22.8 | 19.2 / 23.3 | 19.1 / 22.1 | 19.7 / 24.5 | 19.2 / 23.5 | 20.7 / 24.9 | 19.9 / 23.9 | 19.9 / 24.6 | 20.2 / 23.2 | 20.1 / 24.4 |
| R.m.s.d. d | ||||||||||
| Bonds (Å) / Angles (°) | 0.012 / 1.4 | 0.01 / 1.49 | 0.01 / 1.4 | 0.013 / 1.5 | 0.012 / 1.4 | 0.012 / 1.5 | 0.01 / 1.39 | 0.011 / 1.58 | 0.01 / 1.33 | 0.01 / 1.33 |
| Chiral volume (Å3) | 0.11 | 0.10 | 0.09 | 0.11 | 0.09 | 0.10 | 0.10 | 0.11 | 0.09 | 0.09 |
| Mean B-factors (Å) | ||||||||||
| Main chaine | 43.0 (48.8) | 49.4 (54.3) | 47.6 (52.8) | 52.7 (58.7) | 56.0 (60.8) | 53.8(59.1) | 51.3 (57.9) | 55.5 (61.7) | 52.1 (60.0) | 54.9 (60.1) |
| Side chain | 43.9 (50.3) | 50.2 (55.4) | 48.7 (54.3) | 53.7 (60.3) | 57.4 (62.4) | 54.9 (60.1) | 52.1 (59.2) | 56.6 (63.2) | 52.9 (60.9) | 55.7 (61.2) |
| Solvent / Carbohydrate | 27.2 / - | 32.5 / 80.4 | 30.7 / 80.6 | 32.7 / - | 39.1 / 102.9 | 27.7 / - | 29.7 / - | 32.8 / - | 27.9 / 76.7 | 34.5 / - |
| Ligand / PEG | 43.5 / 42.5 | 55.1 / 65.0 | 57.3 / 91.6 | 60.1 / - | 63.6 / 63.8 | 61.3 / 76.8 | 63.3 / - | 66.9 / 65.9 | 50.6 / 66.1 | 76.0 / 71.0 |
| PDB accession code | 2XUD | 2XUG | 2XUF | 2XUH | 2XUI | 2XUJ | 2XUK | 2XUO | 2XUP | 2XUQ |
Values in parentheses are those for the last shell.
Rsym = Σhkl Σi | Ihkli - <Ihkli>|/ Σhkl Σi <Ihkli>, where I is an individual reflection measurement and <I> is the mean intensity for symmetry-related reflections.
R-factor = Σ ||Fo| – |Fc|| / Σ |Fo|, where Fo and Fc are observed and calculated structure factors, respectively.
Root-mean-square deviation from ideal values.
Values in parentheses are those for subunit B
na, not applicable.
2.2. Structure determination and refinement
The apo mAChE structure (accession code, 1J06)17 without solvent was used as a starting model to refine the structures of the Tyr337Ala mutant and each of its anti1 and syn1 complexes with the program REFMAC21 (Table 2). Rigid-body refinement was performed on each of the two subunits in the crystalline dimer using all data followed by cycles of restrained refinements. A random set of reflections (2%, 2255 reflections) taken from the mAChE-SCh complex structure (2HA2)22 was set aside for cross validation purposes In each case, the resulting sigmaA-weighted 2Fo-Fc and Fo-Fc electron density maps were used to position the inhibitor and correct the protein model with the graphics program COOT23.
The final structures, comprising one Tyr337Ala mutant and its eight co-crystallized and three soaked complexes, comprise residues Glu1-Ala541 and Glu4-Thr540 for the two respective enzyme molecules in the asymmetric unit17. High temperature factors and weak electron densities are associated with the short Ω loop Cys257-Cys272 and the surface loop region Asp491-Pro498. In the Tyr337Ala structure, traces of decamethonium and acetate arising from the purification and crystallization procedures, respectively, are detected in the active site gorge of one subunit in the dimer, as found in previous mAChE structures (accession codes, 1MAA and 1N5R). The stereochemistries of the structures were analyzed with MolProbity24; with the exception of the catalytic Ser203, no residues were found in the disallowed regions of the Ramachandran plot. The rmsd's between the structures of apo mAChE, Tyr337Ala, the anti1- and syn1-mAChE complexes, and each of the anti1- and syn1-Tyr337Ala complexes are in the 0.15-0.32 Å range for 534 Cα atoms. The atomic coordinates and structure factors of the Tyr337Ala structure, the six 1-wk, 1-mth and 10-mth structures of co-crystallized anti1 and syn1 complexes and the three structures of anti1 and syn1 complexes obtained from soaked crystals have been deposited with the Protein Data Bank (cf. Table 2 for accession codes). Figures were generated with PyMOL25.
2.3. Inhibition / binding studies
The Tyr124Gln and Tyr72Asn/Tyr124Gln/Trp286Ala mutants of mAChE were expressed, sequence verified and concentrated from the expression medium as described.15,22 Kinetic analysis of anti1 and syn1 binding to these along with the Tyr337Ala mutants used conventional procedures. The association (kon) and first-order dissociation rate constants (koff) and the equilibrium dissociation constants (Kd) of the anti1 and syn1 isomers for Tyr337Ala were determined as described.11,14 Briefly, association rates were determined by direct stopped-flow measurements of AChE tryptophan quenching upon inhibitor binding, while dissociation rates were determined by spectrophotometric measurements of the time-dependant recovery of AChE activity18 of the AChE-inhibitor complex diluted 5000-fold into a 250 μg/ml solution of herring sperm DNA (Boehringer) or a 50-400 nM solution of the inactive mAChE mutants Ser203Ala22 or Ser203Ala/Tyr337Ala, or a mixture of both DNA and Ser203Ala mutant. Both DNA and mAChE mutants were used as a scavenger to sequester the released inhibitor through its phenanthridinium moiety (DNA) or overall structure (mAChE mutants). All measurements were performed in 0.1 M NaPO4, pH 7.0, at 22°C.
3. Results and discussion
The mAChE mutants Tyr124Gln, Trp286Ala, Tyr337Ala and Tyr72Asn/Tyr124Gln/Trp286Ala were designed previously to mimic tertiary structure of BChE in the PAS domain of mAChE along with, for Tyr337Ala, the choline binding site.15 Mutant Trp286Ala was analyzed for anti1 and syn1 binding in a previous study14 while the other three mutants were analyzed for anti1 and syn1 binding in the present study (Table 1).
3.1. Kinetic analysis of TZ2PA6 isomer binding to the mAChE mutants
Kinetic parameters recorded in solution for anti1 and syn1 binding to the Tyr124Gln mutant led to similar association rates, and hence, comparable dissociation constants as for mAChE (Table 1). For the Trp286Ala and triple mutants, 5-fold to 340-fold greater dissociation constants dictated by up to 30-fold slower association and 10-fold slower dissociation were recorded, consistent with involvement of these residues that line the active site gorge in stabilization of either of the anti- and syn-triazoles in mAChE. For the mutants, particularly the triple mutant, conformationally distinct complexes in the gorge confines can thus be expected from removal of large aromatic side chains.
In contrast, for the Tyr337Ala mutant compared to mAChE, similar association rates but much lower dissociation rates, by 3000-fold for the anti1 and 550-fold for the syn1, were recorded, whereas they were lowered by only 350-fold and 85-fold when compared with T. californica AChE, with its Phe substitution for Tyr337 (Table 1). The resulting femtomolar affinities largely reflect greater activation barriers, and place the TZPA isomers at a similar free energy of binding as that of 1,1,1-trifluoroacetophenone derivatives towards AChE or biotin towards streptavidin, despite a higher number of non-hydrogen atoms (Supporting Information, Fig. S1).26 Compared with the anti1 isomer, the 4-fold greater affinity of the syn1 isomer for the Tyr337Ala mutant also arises from a 5-fold lower dissociation rate. This results in half-lives in solution of more than one month (t1/2: 37 days) for the syn1 complex and one week (t1/2: 7.3 days) for the anti1 complex, compared with t1/2 values of 98 min and 3 min for the respective mAChE complexes. This also suggests that in the crystallization set-up, where dissociation is largely precluded by the high protein concentration of the complexes (107-109-fold the Kd in the mother liquor, presumably ≥ 1012-fold the Kd in the crystal), equilibrium would be largely displaced toward complex formation, resulting in even higher stability of the complexes.
To explore this issue, for each of the co-crystallized anti1- and syn1-Tyr337Ala complexes we collected complementary data sets at increasing times of crystal maturation corresponding to smaller, near or greater the half-time values for complex dissociation in solution (Table 2). Another set of complexes, obtained by soaking Tyr337Ala crystals with individual or mixed inhibitors for times much shorter than the solution half-lives of the respective complexes (cf. Section 2.1.), was also analyzed (Table 2).
3.2. Overall fold of the Tyr337Ala mutant
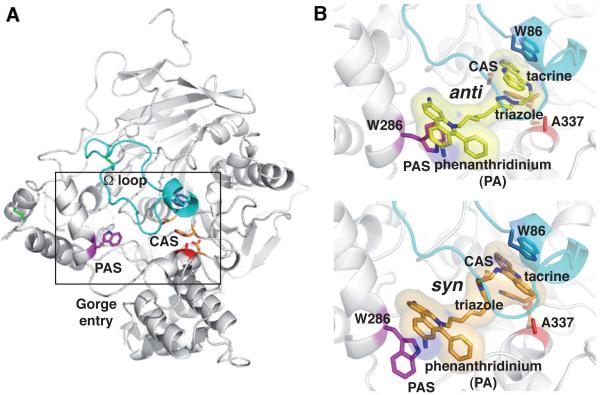
The structure of the Tyr337Ala mutant is identical to that of mAChE (rmsd 0.28 Å for 535 Ca atoms), except at the substitution position where replacement of the bulky Tyr phenol ring by the small Ala methyl group enlarges, by ~2.5 Å, the gorge internal diameter (Fig. 1). Yet at this position the deviation for the gorge walls is only ~0.5 Å, indicating that the gorge shape is virtually unaltered and supporting catalytic activity similar to mAChE (data not shown). For each of the anti1- and syn1-Tyr337Ala complexes, 1-wk, 1-mth and 10-mth structures were solved (Table 2). In all six structures, the bound inhibitors are well-ordered and their binding site may be deconstructed into the same three discrete loci as found for the anti1- and syn1-mAChE complexes: i) the active center at the base of the gorge where the tacrine moiety binds; ii) an intervening site in the constricted region within the gorge that associates with the triazole moiety and adjacent methylene groups; iii) the PAS at the gorge rim where the phenylphenanthridinium moiety binds (Fig. 1).14 However, significant differences in the conformations of the inhibitor molecules and the positions of side chains within the gorge are observed between each pair (anti1 versus syn1) of the TZ2PA6-Tyr337Ala complexes.
Figure 1. Overall views of the Tyr337Ala subunit and bound TZ2PA6 inhibitors.
(A) Overall view of the mAChE-Tyr337Ala molecule (white ribbon), viewed down into the gorge. The catalytic triad residues Ser203, Glu334, His447 are displayed in orange at the center of the subunit. The long Ω loop Cys69-Cys96 is in cyan, PAS residue Trp286 is in magenta, the substituted Ala337 in red and Trp86 at the base of the gorge in blue. The three disulfide bridges respectively located in the long Ω loop, the short Ω loop (Cys257-Cys272) and between helices α(4)7,8 and α10 in the four-helix bundle are displayed as green bonds. (B) Close-up views of the bound anti1 (top, yellow bonds and transparent molecular surface) and syn1 isomers (bottom, orange bonds and surface) and surrounding side chains in the 1-mth structures (same orientation as in panel A).
3.3. The three anti1-Tyr337Ala complexes obtained by co-crystallization
The binding of the anti1 isomer to Tyr337Ala in the 1-wk structure (corresponding to 1-fold the t1/2 for anti1) drastically differs from its binding to mAChE (Figs. 2 and 3). At the bottom of the gorge, the tacrine moiety, although positioned similarly as in mAChE, is flipped upside-down and tilted, by ~25°, compared to its position in the anti1- and syn1-mAChE complexes. As a result, the polar interaction that, in the anti1- and syn1-mAChE complexes, occurs between the basal tacrine N8 atom and the backbone carbonyl of His447, is now swapped to involve the opposite N7 atom at the basis of the dimethylene linker connecting the tacrine and triazole.
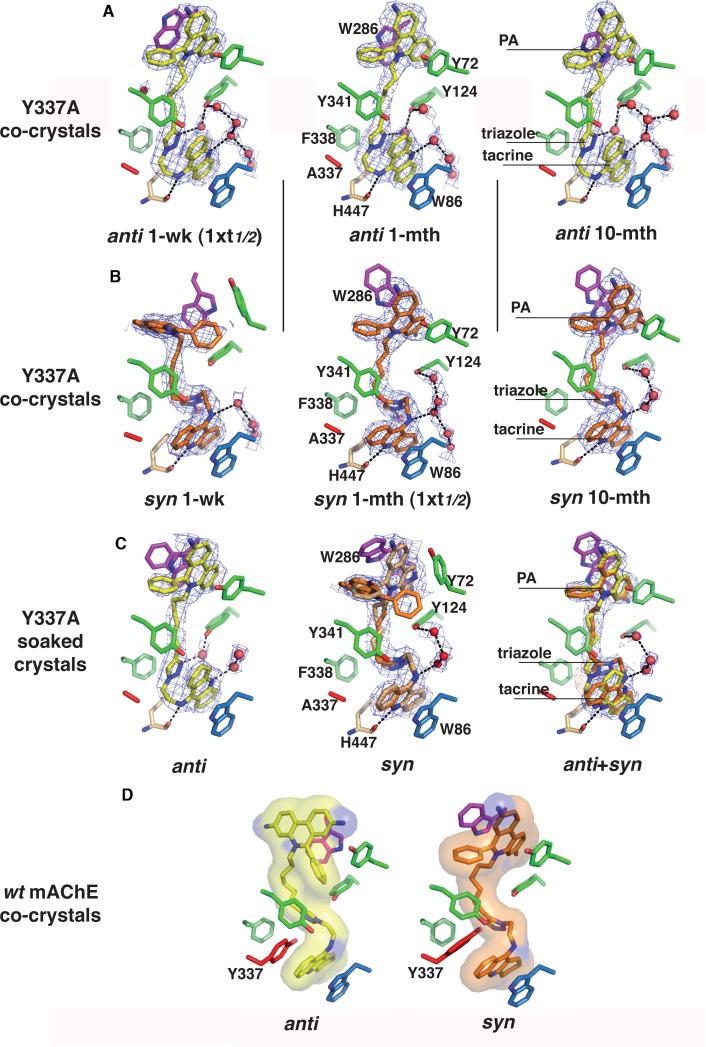
Figure 2. Close up views of the TZ2PA6-Tyr337Ala complexes.
Bound anti1 (A, yellow) and syn1 (B, orange) in the 1-wk, 1-mth and 10-mth structures (left to right) oriented as in Fig. 1B. The respective 2.55-2.8 Å resolution final 2Fo-Fc electron density maps are contoured at 1 σ (cyan). The key interacting side chains are colored blue, green and magenta (blue nitrogens, red oxygens) for those that respectively interact with the tacrine, triazole and phenanthridinium moieties of the isomers. The catalytic His447 is displayed in light orange and the side chain of Ala337 is in red. Water molecules mediating ligand interactions are shown as red spheres. Hydrogen bonds between mAChE-Tyr337Ala, the bound isomers and the water molecules are shown as dotted lines. (C) Bound anti1 (left, yellow), syn1 (middle, orange) and anti1/syn1 (right, yellow and orange, respectively) in the structures obtained by crystal soaking. The soaked syn1 when bound adopts two conformations. Soaking an anti1/syn1 mixture leads to a predominant syn1 and a low abundance anti1, as demonstrated by the positive peaks above 3.5 σ (red) in the difference density maps around the triazole moiety. (D) The anti1 and syn1 isomers as bound to mAChE (PDB codes, 1Q84 and 1Q83)14. The side chain of Tyr337 is shown in red. The molecular surfaces of the isomers are displayed in transparency.
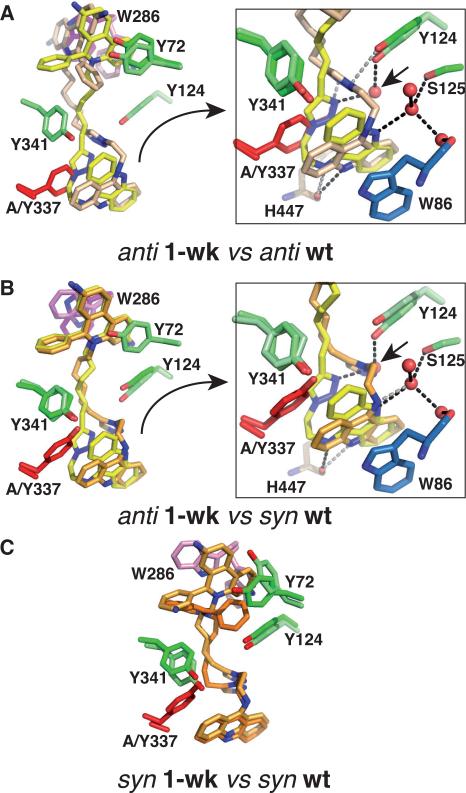
Figure 3. Comparison of the most divergent Tyr337Ala and mAChE complexes.
(A) Overall view (left) and close up view (right) of the tacrine and triazole binding regions of overlayed anti1 in the 1-wk structure (yellow bonds) and anti1 bound to mAChE (light orange bonds). (B) Overall view (left) and close up view (right) of the tacrine and triazole binding region of superimposed anti1 in the 1-wk structure (yellow bonds) and syn1 bound to mAChE (light orange bonds). (C) Overall view of superimposed syn1 in the 1-wk structure (orange bonds) and syn1 in mAChE (light orange bonds). The side chains of Trp286 and of Tyr72, Tyr124 and Tyr341, which adopt distinctive positions in the complexes, are displayed in magenta/pink and in green/light green, respectively. The side chain of Ala/Tyr337 is in red and that of Trp86 in blue. The short arrow in the framed figures points to a water molecule, in the 1-wk structure of the anti1-Tyr337Ala complex, that mimics the triazole N2 nitrogen atom in the syn1-mAChE complex. Superimpositions were made according to all Cα atoms in the Tyr337Ala or mAChE subunit.
At the region of constriction formed by the side chains of Tyr124, Phe297 and Phe338 some 5-8 Å into the gorge, where the substituting Ala337 side chain is located, the hooked conformation of the dimethylene chain dictates a distinct positioning for the triazole, which in the Tyr337Ala mutant is displaced by 3 Å laterally into the gorge and oriented ~90° away from its position in mAChE, as to mimic the tip of the missing Tyr337 phenol ring (Figs. 2 and 3). As a result, the triazole establishes stacking interactions in near-sandwich conformation between the central 6-heterocycle ring of tacrine and the Phe338 phenyl rings. In turn, the first methylene of the hexamethylene linker that connects the triazole and phenylphenanthridinium is positioned 2.3 Å deeper into the gorge and adopts an extended, linear conformation instead of the curved shape observed in the anti1-mAChE complex. The large differences in the triazole and hexamethylene positions in the Tyr337Ala mutant compared to mAChE are associated with discrete positional rearrangement of the Tyr124 and Tyr341 side chains that line the gorge wall.
At the PAS, the phenanthridinium intercalates between the Tyr72 and Trp286 side chains while the Trp286 side chain, dislodged from the PAS surface, adopts a solvent-exposed position similar to its position in the syn1-mAChE complex (Fig. 2). However, compared to this complex, a ~180° flip of the Trp286 side chain now induces a different, albeit still parallel, stacking interaction with the phenanthridinium. In fact, the intercalated phenanthridinium stabilizes an alternate minor conformation of the Trp286 side chain, flipped by 150°, reminiscent of that observed for the corresponding Trp279 residue in the TcAChE-NF595 complex27. The mode of binding of the phenanthridinium ring at the PAS, that drastically differs from that observed in the anti1-mAChE complex, results in a deeper positioning of the triazole into the gorge associated with an extended conformation of the hexamethylene linker.
Analysis of the anti1-Tyr337Ala complex in the 1-mth structure (4-fold the t1/2) compared to the 1-wk structure reveals no significant differences in the isomer and enzyme conformations at the bottom of the gorge and the mid-gorge region of constriction (Fig. 2). Yet at the PAS, the Trp286 side chain, dislodged from the PAS surface, is positioned mid-way the two alternate positions seen in the 1-wk structure with no significant difference in the orientation of the phenanthridinium.
In the anti1-Tyr337Ala complex in the 10-mth structure (42-fold the t1/2), the isomer and enzyme conformations are similar as in the 1-mth structure, whereas the Trp286 side chain adopts a similar conformation as the minor alternate conformation observed in the 1-wk structure (Fig. 2). Hence even in the crystalline enzyme, the Trp286 side chain retains sufficient flexibility upon binding of a extremely potent ligand to undergo multiple fluctuating orientations, here exemplified by a 50° shutter-like movement of its indole ring in the 1-wk and 10-mth structures.
3.4. The three syn1-Tyr337Ala complexes obtained by co-crystallization
The binding of the syn1 isomer to the Tyr337Ala mutant in the 1-wk structure (0.2-fold the t1/2 for syn1) also differs from its binding to mAChE, besides a similarly oriented tacrine at the base of the gorge (Figs. 2 and 3). At the region of constriction, the triazole is shifted (by 1 Å) deeper into the gorge where it undergoes a near-parallel stacking interaction with the tacrine central amino-substituted ring. In turn, the hexamethylene linker is also shifted down (by ~1.5 Å) into the gorge, a feature that resembles the differences seen in the positions of the linker between the anti1 and syn1 isomers bound to mAChE14.
At the PAS, the slightly deeper position of the triazole causes the syn1 phenylphenanthridinium to adopt yet another distinct conformation, not observed in either structures of the anti1-Tyr337Ala or anti1- and syn1-mAChE complexes (Figs. 2 and 3). The phenanthridinium, here oriented perpendicular to the gorge axis with one face buried some 2 Å deep into the gorge and the other face exposed to the solvent, totally occludes the gorge entry. One of the distal ring is anchored to the Leu289-Arg296 loop region, with the nitrogen hydrogen bonded to the Ser293 and Arg296 carbonyl atoms, while the opposite distal ring weakly establishes an edge-to-face interaction with the Tyr341 phenol ring. The exocyclic phenyl ring in the phenanthridium inserts between the Tyr341 and Trp286 side chains, while the Trp286 side chain, in the typical orientation of apo mAChE, establishes van der Waals interactions with the phenyl ring. The newly positioned phenylphenanthridinium induces a ~40° arc motion of the Tyr72 phenol ring and a 30° tilt of the Tyr341 phenyl ring, respectively located on each side of the gorge entrance compared to their respective positions in the syn1-mAChE structure. In fact, the final density maps reveal additional electron density in the vicinity of the phenyl ring that could correspond to an alternate, minor abundance conformation of the syn1 phenanthridinium, not observed in the other anti1- or syn1-Tyr337Ala complex structures.
Analysis of the syn1-Tyr337Ala complex in the 1-mth structure (0.8-fold the t1/2) reveals, for the bound tacrine and triazole at the base of the gorge and the hexamethylene linker at the region of constriction, similar positions and orientations as in the 1-wk structure (Fig. 2). At the PAS however, the phenanthridinium ring is oriented ~115° away from its position in the 1-wk structure as to match the orientation previously observed in the syn1-mAChE complex (Fig. 3). Consequently, the Trp286 side chain is dislodged from the PAS surface and swings towards the solvent to adopt a conformation that will be retained in the 10-mth structure (8-fold the t1/2) (Fig. 2). Hence, in two distinctively matured crystals grown from the same syn1-Tyr337A complex, two distinct conformations of the phenanthridinium ring were trapped at the PAS. Moreover, the ligand pose and enzyme conformation observed in the 10-mth structure are essentially retained in even older complexes (cf. Section 2.1.), consistent with a thermodynamic equilibrium being attained at maturation times matching the solution half-lives.
These comparative data, combined with the structures of mAChE bound with the same inhibitors, demonstrate that albeit a femtomolar affinity, distinct conformations of the inhibitor coupled to conformational adaptations at the PAS exist, some of minor abundance, and thus reflect the behavior of the enzyme in solution. This is well exemplified by the presence of two alternate conformations of the Trp286 side chain in the syn1-Tyr337Ala complex in the 10-mth structure, of which one is similar to that in the 1-mth structure (Fig. 2).
3.5. The anti1- and syn1-Tyr337Ala complexes obtained by crystal soaking
Compared with the anti1-Tyr337Ala structures obtained by co-crystallization, that obtained from a crystal soaked for a time 7-fold shorter than the complex half-life (Table 2; cf. Section 2.1.) shows a similar mode of binding for the anti1 isomer as found in the 1-wk structure, with only slight variations in the hexamethylene linker conformation and the Trp286 side chain orientation at the PAS (Fig. 2). In contrast, compared with the syn1-Tyr337Ala structures obtained by co-crystallization, that obtained from a soaked crystal (here for a time 30-fold shorter than the complex half-life (Table 2)) reveals two alternate conformations of the phenanthridinium ring at the PAS, of which each matches the orientation respectively identified in the syn1-Tyr337Ala 1-wk and 1-mth structures (Fig. 3). Hence, crystal soaking procedures led to picturing freshly formed complexes resembling more the complexes observed at crystal maturation times shorter than the solution half-lives.
In turn, the structure derived from soaking an equimolar anti1/syn1 mixture reveals that the syn1 isomer was predominantly trapped in the active site gorge, the anti1 isomer being present in lower abundance only (Fig. 2). This is consistent with the kinetic data showing the higher affinity of the syn1 isomer to reflect at least a 10-fold lower dissociation rate. Hence, either isomer can occupy the active site gorge at the same rate, but the syn1 complex, owing to its slower dissociation rate, prevents anti1 from binding again. Moreover, in this complex the bound syn1 adopts the most energetically favorable conformation observed in the mature syn1-Tyr337Ala 1-mth and 10-mth structures. Hence, “dynamic combinatorial X-ray crystallography” (or DCX) on a limited scale can be used to select, from a mixture of structurally related high-affinity inhibitors, the slowest dissociating one. This observation, along with the reported binding selectivity of crystalline TcAChE for the soaked (R)- versus (S)-enantiomers, with otherwise similar affinities, of bifunctional inhibitors E22028 and tacrine-(10)-hypyridone29, argues for a structure-based approach to conduct inhibitor discovery and optimization.
3.6. Implications of structure variation
Comparison of the structures obtained for complexes of mAChE and its Tyr337Ala mutant with the TZ2PA6 anti1 and syn1 regioisomers emphasizes the key role of the triazole located midway the gorge in dictating the orientation of the phenanthridinium at the PAS. This is consistent with our previous proposal that the 1.5 Å positional shift between the anti1 and syn1 triazoles within the mAChE gorge influences the orientation of the phenanthridinium ring. Here we show that in the absence of the protruding Tyr337 side chain, the anti1 triazole adopts a deeper position within the gorge of the mutant, roughly equivalent to that observed for the syn1 isomer bound to mAChE and resulting in a similar orientation of the intercalated phenanthridinium ring. Similarly, the deeper positioning of the syn1 triazole in the Tyr337Ala mutant compared to its position in mAChE results in a new orientation of the phenanthridinium ring, which seems to be fluctuating between two distinct orientations as exemplified in the 1-wk and 1-mth structures (Fig. 2).
The Tyr337Ala mutant is the only one of our screened mutants where the affinities of both the syn1 and anti1 complexes increase rather than decrease over the wild-type enzyme, a feature that suggests that the presence of the aromatic ring places some steric constraints around the triazole. Indeed, mutation of the facing Tyr124 only moderately affects the binding affinitiy of the two inhibitors, consistent with the structures of the complexes (Table 1). Since the ligands bind at near the diffusion rate limitation, the enhanced affinity is reflected in a slower dissociation rate.
A comparison of the multiple structures points to the large differences in the overall conformation of the inhibitor bound to the mutant. Importantly, these conformations are not specific to the crystalline phase, but rather could reflect the fact that reactants, when simultaneously bound, induce these minor abundance conformational changes. Indeed, a combination of flexibility of reactant chain length and subtle rearrangements of the key interacting side chains are required for the 1,3-dipolar cycloaddition reaction to occur. Therefore the conformational fluctuations seen in the trapped inhibitor could reflect the conformational behavior of reactants. Decay of fluorescence anisotropy experiments along with an overlay of all available mAChE crystal structures in complex with ligands or conjugates argue towards this point (Z. Radic, unpublished data).
However, what remains obscure is whether the freeze frame inhibitor syn1, initially tailored in the AChE active site gorge and formed in situ11, comes from a minor abundance complex of the reactants and its formation is driven by proximity of the reactive azide or alkyne, or if reactant binding itself selects a dominant conformation. Our data highlight the dynamic behavior of the AChE gorge, most particularly near its rim, and are consistent with the concept of protein breathing motions that are altered by the reactants. In the crystal, local packing forces do not modify the thermodynamic energy balance, selecting different conformations. Indeed, whereas the rigid anti1 isomer adopts an elongated conformation within the gorge and has to fold into a hook in the catalytic pocket and rotate its top and bottom moieties in opposite directions (like a classic pepper mill) to become shorter, the highly flexible syn1 isomer adopts a compacted and torturous conformation that can easily be compressed or extended (like a spring), depending on the position of the triazole into the gorge that influences the orientation of the phenantridinium at the PAS. The modes of interaction between the isomers and the Tyr337Ala mutant of mAChE, dominated by apolar interactions with contribution of conserved bridging water molecules, are similar for each of the six co-crystallized complexes, indicating that the overall shape rather than a particular mode of interaction might be a predominant factor for this gain in affinity. At the PAS, the intercalation of the phenylphenanthridinium between the Trp286 and Tyr72 side chains, so far only seen for the syn1 isomer bound to mAChE14 and now extended to the anti1 isomer bound to Tyr337Ala, could significantly contributes to the large gain in affinity. Indeed, this peculiar conformation at the PAS could favor a near parallel rather than an antiparallel orientation of the azide and alkyne reactants (syn1- over anti1), causing the bond distance between the tacrine and phenanthridium moieties to be foreshortened by ~1.2 - 1.4 Å.
4. Conclusions
The conformational and positional differences observed throughout the four structures of the anti1-Tyr337Ala complex on one hand and four structures of the syn1-Tyr337Ala complex on the other hand (one soaked and three co-crystallized complexes in each case) appear to follow the increasing ratio of maturation time to dissociation half-time and hence, the time required for optimal stabilization of the respective crystalline complexes. These differences are particularly critical for the phenylphenanthridinium/hexamethylene portion of the inhibitors and surrounding enzyme residues in the upper part of the gorge and at the PAS, whereas the triazole/dimethylene/tacrine portion and surrounding residues at the bottom of the gorge adopt a definitive conformation and position perhaps as soon as each complex forms. These observations are consistent with the requirement for a more rigid active center to insure substrate specificity and rapid hydrolysis, while a more flexible PAS and gorge rim offer a wide range of recognition and binding capacities for allosteric regulation of catalysis.
The high affinities of the syn1- and anti1-Tyr337Ala complexes, with their dissociation times exceeding those of nucleation and crystal growth, afford a unique opportunity to examine conformational states dictated by ligand association, a kinetically dominated process, and those that reflect attainment of thermodynamic equilibrium, achieved by slow isomerization of the complex and/or multiple association and dissociation steps leading to the slowest dissociating ligand becoming the dominantly bound one. A critical question is the conversion from the kinetically driven ligand poses to those found at longer times of equilibrium. Free ligand concentrations are high in the soaking experiments, but they are extremely low when co-crystals are formed at a stoichiometry of one ligand per active site. Hence, for the co-crystals one can exclude unbound ligand in the bulk solvent participating in the conversion in conformational states. This leaves two possibilities: (i) the conversion occurs in situ in the crystal analogous to a unimolecular isomerization; (ii) dissociation occurs into the limited space of the crystal solvent channels and then reassociation ensues, allowing the more slowly dissociating complexes to become dominant upon time. Since an isomerization of the complex and the dissociation process would both require Boltzman energy, they may be kinetically indistinguishable without assessing the presence of free ligand in the system. However, the very high protein concentrations in the crystal and surrounding solution (cf. Section 3.1.) along with the low temperature used for crystal formation and maturation (cf. Section 2.1.) most likely preclude complex dissociation. This suggests in cristallo isomerization to be the main conversion process from the kinetically driven complexes observed from the younger crystals to the thermodynamically driven equilibrium observed from the older crystals.
Distinctive large- or small-scale conformational changes of part or all of a crystalline receptor or enzyme have been evidenced by comparing structures solved from distinct crystal forms, crystallization conditions, complexes or any combination of these.14,30-34 Alternative conformations of subunits within a same oligomeric protein have been reported,34,35 and alternative conformations or binding positions of a bound ligand relative to its binding site have been described or predicted.17,36,37 Yet, to our knowledge our study is a first to report a range of alternative conformations for both the ligand and protein in “same litter” crystalline complexes equilibrated over time periods consistent with or greater than their half-lives. Since kinetic and thermodynamic rules governing complex remodeling can still occur in the crystalline state, and since multiple poses of bound ligands of near equivalent free energy can be found in the structures, our data also suggest that statement “the crystal structure” of a ligand-protein complex should be interpreted as “a crystal structure” (i.e., perhaps amongst several possible) of this complex.
Supplementary Material
Acknowledgements
We are grateful to M. Juin and S. Conrod (CRN2M, Marseille) for assistance in crystallization and crystal soaking; to G. Sulzenbacher, S. Spinelli and M. Czjzek, (AFMB, Marseille) and the ID14 and ID29 staff of the European Synchrotron Radiation Facility (ESRF, Grenoble) for expert contribution to data collection; and to K.B. Sharpless and H.C. Kolb (Dept of Chemistry and Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA) for the kind gift of the TZ2PA6 regioisomers, critical reading of the manuscript and fruitful discussions. This work was supported by grants from the USPHS and DAMD (to PT) and the Association Française contre les Myopathies and the CNRS-Direction des Relations Internationales (to PM).
Abbreviations
- AChE
acetylcholinesterase (mAChE, recombinant from mouse)
- PAS
peripheral anionic site
- syn1-TZ2PA6
3,8-diamino-6-phenyl-5-[6-[1-[2-[(1,2,3,4-tetrahydro-9-acridinyl)amino]ethyl]-1H-1,2,3-triazol-5syn-yl]hexyl]-phenanthridinium (C42H45N8)
- anti1-TZ2PA6
3,8-diamino-6-phenyl-5-[6-[1-[2-[(1,2,3,4-tetrahydro-9-acridinyl)amino]ethyl]-1H-1,2,3-triazol-4anti-yl]hexyl]-phenanthridinium (C42H45N8)
Footnotes
Supporting Information Available: Supplementary Figure S1 and associated references. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Quinn DM. Chem. Rev. 1987;87:955–79. [Google Scholar]
- 2.Taylor P. Neurology. 1998;51:S30–35. discussion S65-67. Review. [Google Scholar]
- 3.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–9. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 4.Taylor P, Lappi S. Biochemistry. 1975;14:1989–97. doi: 10.1021/bi00680a029. [DOI] [PubMed] [Google Scholar]
- 5.Greenblatt HM, Guillou C, Guénard D, Argaman A, Botti S, Badet B, Thal C, Silman I, Sussman JL. J. Am. Chem. Soc. 2004;126:15405–11. doi: 10.1021/ja0466154. [DOI] [PubMed] [Google Scholar]
- 6.Berman HA, Decker MM, Nowak MW, Leonard KJ, McCauley M, Baker WM, Taylor P. Mol. Pharmacol. 1987;31:610–6. [PubMed] [Google Scholar]
- 7.Haviv H, Wong DM, Silman I, Sussman JL. Curr. Top. Med. Chem. 2007;7:375–387. doi: 10.2174/156802607779941215. [DOI] [PubMed] [Google Scholar]
- 8.Kolb HC, Sharpless KB. Drug Discov Today. 2003;8:1128–37. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 9.Tron GC, Pirali T, Billington RA, Canonico PL, Sorba G, Genazzani AA. Med. Res. Rev. 2008;28:278–308. doi: 10.1002/med.20107. [DOI] [PubMed] [Google Scholar]
- 10.Moorhouse AD, Haider S, Gunaratnam M, Munnur D, Neidle S, Moses JE. Mol. Biosyst. 2008;4:629–42. doi: 10.1039/b801822g. [DOI] [PubMed] [Google Scholar]
- 11.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Angew Chem. Int. Ed. 2002;41:1053–7. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Manetsch R, Krasinski A, Radic Z, Raushel J, Taylor P, Sharpless KB, Kolb HC. J. Am. Chem. Soc. 2004;126:12809–18. doi: 10.1021/ja046382g. [DOI] [PubMed] [Google Scholar]
- 13.Krasinski A, Radic Z, Manetsch R, Raushel J, Taylor P, Sharpless KB, Kolb HC. J. Am. Chem. Soc. 2005;127:6686–92. doi: 10.1021/ja043031t. [DOI] [PubMed] [Google Scholar]
- 14.Bourne Y, Kolb HC, Radic Z, Sharpless KB, Taylor P, Marchot P. Proc. Nat. Acad. Sci. USA. 2004;101:1449–54. doi: 10.1073/pnas.0308206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radic Z, Pickering NA, Vellom DC, Camp S, Taylor P. Biochemistry. 1993;32:12074–84. doi: 10.1021/bi00096a018. [DOI] [PubMed] [Google Scholar]
- 16.Marchot P, Ravelli RBG, Raves ML, Bourne Y, Vellom DC, Kanter J, Camp S, Sussman JL, Taylor P. Protein Sci. 1996;5:672–9. doi: 10.1002/pro.5560050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourne Y, Taylor P, Radic Z, Marchot P. EMBO J. 2003;22:1–12. doi: 10.1093/emboj/cdg005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellman GL, Courtney KD, Andres V, Jr., Featherstone RM. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.CCP4, Collaborative Computational Project Number 4 Acta Crystallogr. 1994;D50:760–3. [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. 1997;D53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Bourne Y, Radic Z, Sulzenbacher G, Kim E, Taylor P, Marchot P. J. Biol. Chem. 2006;281:29256–67. doi: 10.1074/jbc.M603018200. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Acta Crystallogr. 2004;D60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLano WL. The PyMOL Molecular Graphics System. Delano Scientific; San Carlos, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
- 26.Kuntz ID, Chen K, Sharp KA, Kollman PA. Proc. Natl. Acad. Sci. USA. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colletier JP, Sanson B, Nachon F, Gabellieri E, Fattorusso C, Campiani G, Weik M. J. Am. Chem. Soc. 2006;128:4526–7. doi: 10.1021/ja058683b. [DOI] [PubMed] [Google Scholar]
- 28.Kryger G, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with E2020 (Aricept®): implications for the design of new anti-Alzheimer drugs. Structure. 1999;7:297–307. doi: 10.1016/s0969-2126(99)80040-9. [DOI] [PubMed] [Google Scholar]
- 29.Haviv H, Wong DM, Greenblatt HM, Carlier PR, Pang Y-P, Silman I, Sussman JL. Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase. J. Am. Chem. Soc. 2005;127:11029–36. doi: 10.1021/ja051765f. [DOI] [PubMed] [Google Scholar]
- 30.Grochulski P, Li Y, Schrag JD, Bouthillier F, Smith P, Harrison D, Rubin B, Cygler M. J. Biol. Chem. 1993;268:12843–7. [PubMed] [Google Scholar]
- 31.Grochulski P, Li Y, Schrag JD, Cygler M. Protein Sci. 1994;3:82–91. doi: 10.1002/pro.5560030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka T, Ames JB, Harvey TS, Stryer L, Ikura M. Nature. 1995;376:444–7. doi: 10.1038/376444a0. [DOI] [PubMed] [Google Scholar]
- 33.Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 34.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. EMBO J. 2005;24:3635–46. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hibbs RE, Sulzenbacher G, Shi J, Talley TT, Conrod S, Kem WR, Taylor P, Marchot P, Bourne Y. EMBO J. 2009;28:3040–51. doi: 10.1038/emboj.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao F, Bern N, Little A, Wang HL, Hansen SB, Talley TT, Taylor P, Sine SM. J. Biol. Chem. 2003;278:23020–6. doi: 10.1074/jbc.M301151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang HL, Gao F, Bren N, Sine SM. J. Biol. Chem. 2003;278:32284–91. doi: 10.1074/jbc.M304366200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.