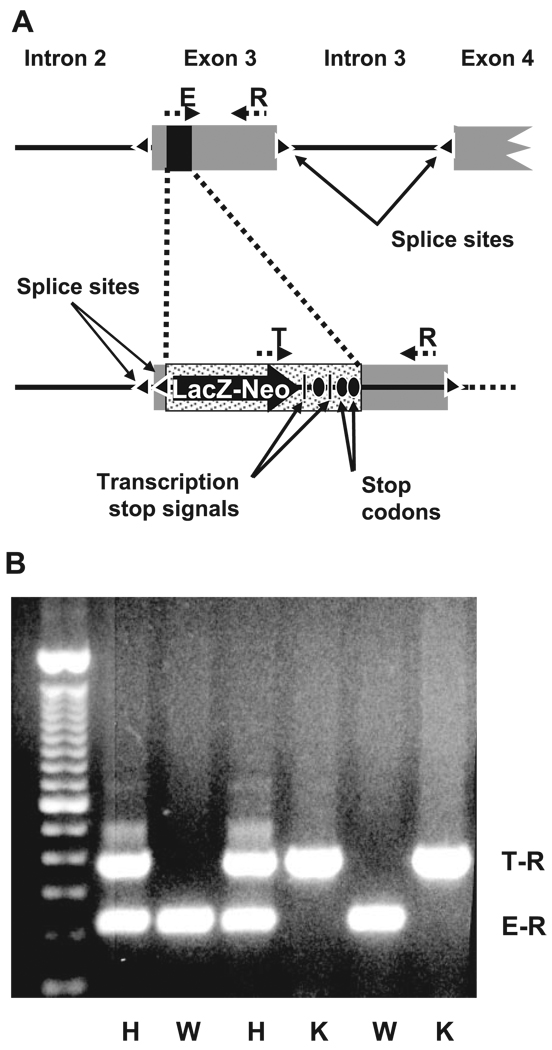
Fig. 1.
Knockout of RST. A: schematic of targeted disruption of RST. Homologous recombination was performed to replace a 70-base segment (black rectangle) in exon 3 of RST with a LacZ-Neo cassette flanked by an upstream splice acceptor site (triangle) and multiple downstream transcription termination signals (vertical black bars) and in-frame stop codons (black ovals), resulting in formation of a null allele. The positions of the genotyping primers R, E, and T are indicated. B: genotyping of a typical litter from a cross between mice heterozygous for the RST null allele. A common “reverse” primer (R in A) and “forward” primers specific to either the endogenous or targeted (null) alleles (E and T, respectively, in A) were used in multiplex PCR on genomic DNA from individual offspring. Primers E and R amplify a 224-bp product (E-R) from the wild-type (WT) allele, while primers T and R amplify a 366-bp product (T-R) from the knockout (KO) allele; thus offspring forming only the E-R product are wild types (indicated by W), those forming only the T-R product are knockouts (K), and those forming both products are heterozygotes (H).