Fig. 3.
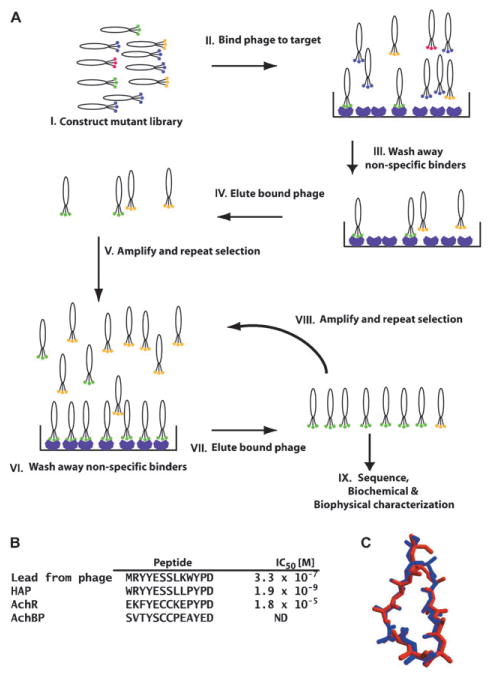
A, schematic of the in vitro selection cycle using phage-display. I: depicts a collection of phage variants in which the library of peptides or proteins is displayed as a fusion to a phage coat protein. Colors indicate individual variants. II: the phage library is mixed with an immobilized purified target protein. III: phages that do not bind are washed away. Some sequences that bind non-specifically, indicated by the yellow hexagons, may remain. IV: recovery of bound phage by elution with ligand or low pH. V: eluted phages are amplified by passage through E. coli. The amplified library of recovered variants is then used in a second round of selection, steps VI through VIII. IX: progress of the experiment is usually monitored by sequencing some fraction of the selected clones. As the cycles of selection progress, the sequence variation of the library should decrease. Once the rounds of selection are finished (generally three–ten rounds), the selected peptide or protein product is made and characterized. In outline, the depicted selection cycle is similar to the procedures used for in vitro selection of nucleic acid aptamers by SELEX. B, left, comparison of α-BXT binding peptides discovered using phage display and subsequent design (HAP) with the sequences of the binding site from the channel (AChR) and AChBP. IC50 values for blocking α-BXT binding to AChR. C, comparison of the structure of the backbone and Cβ positions of the HAP peptide from the HAP–α-BXT complex (red) and AChBP residues (blue). Panels B and C are adapted from ref. 74.
