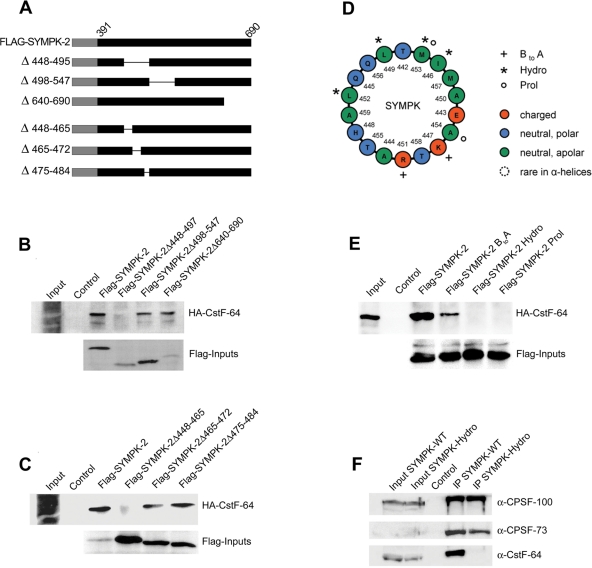
FIGURE 3:
Further characterization of CstF-64 interaction site in symplekin. (A) Schematic drawing of Flag-SYMPK-2 fusion proteins containing deletions. Numbers and a thin line indicate the deleted amino acid residues. (B–C) Mapping of the interaction with CstF-64 by using internal and C-terminal deletions of the symplekin fragment SYMPK-2. Full-length, Flag-tagged SYMPK-2 (corresponding to residues 391–690) and deletions thereof were translated in vitro, incubated with α-Flag M2 agarose, and then incubated with in vitro–translated HA-CstF-64. After precipitation and Western blot, HA-CstF-64 binding was detected with α-HA-HRP. Inputs of the bait proteins were detected by using α-Flag M2-peroxidase. All inputs correspond to 1/10 of the amount used in the pull-down. This analysis defined the CstF-64 interaction region between symplekin residues 391–448 (upstream) and ∼465 (downstream). (D) Helical wheel projection of an α-helix spanning residues 442–460 whose role was further explored with mutants. The projection was generated with freeware developed by Marcel Turcotte at the University of Ottawa, Ontario, Canada and graphically edited. The last amino acid of the helix, A460, could not be rendered by this graphical procedure. The mutated residues are indicated by plus signs (BtoA), asterisks (Hydro), or small circles (Prol; see main text). (E) Analysis of mutant symplekin fragment SYMPK-2 by the binding assay already used in (B) and (C). (F) Coimmunoprecipitation of 3′-end processing proteins with either full-length, C-terminally Flag-tagged symplekin or the identically tagged symplekin Hydro mutant from HeLa total cell extract. Inputs correspond to 1/10 of the material used for the coimmunoprecipitations. Untransfected cells served as negative control.