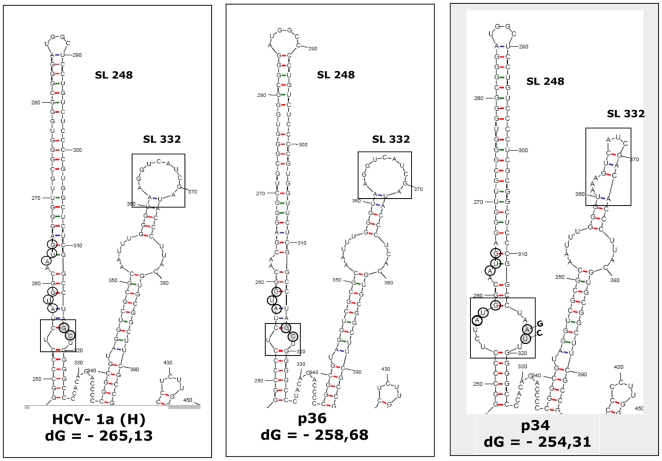
Figure 9. Prediction of stem-loop 248 and stem-loop 332 RNA folds, for HCV-1a H strain, the p36 isolate (positive for anti-core and negative for anti-core+1/ARFP antibodies) and p34 (negative for anti-core and positive for anti-core+1/ARFP).
RNA secondary structure was predicted using the m fold program. Open boxes indicate structure alterations in the internal loop of stem-loop 248 and in the apical loop of stem-loop 332. Open circles represent the locations of internal translation initiation codons (AUG 85 and 87) that serve for core+1/ARFP synthesis. Gray circles represent the nucleotides that are altered in isolates from anti-core+1/ARFP antibody positive patients.