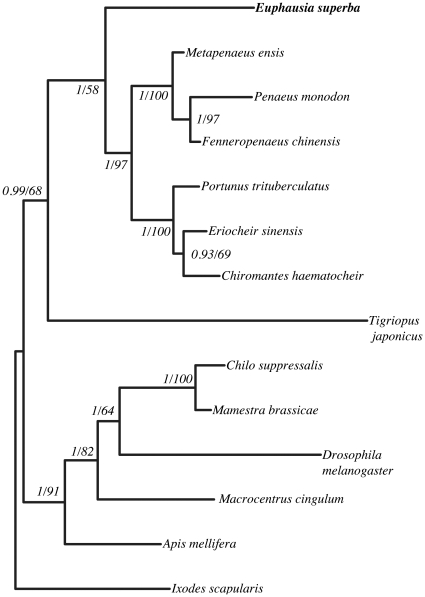
Figure 2. Phylogeny based on Bayesian analysis and maximum likehood of the amino acid data set of HSP90 sequences from the pancrustaceans (14 taxa, 689 characters).
Ixodes scapularis was assigned as outgroup. Trees obtained by Bayesian analysis and by maximum likelihood analysis of the amino acid dataset were fully congruent. Numbers at nodes are posterior probabilities and bootstrap values (based on 100 replicates) respectively obtained from the analysis of the amino acid dataset. Sequence accession numbers are: Ixodes scapularis: XM_002414763; Apis mellifera: FJ713701; Macrocentrus cingulum: EU570066; Drosophila melanogaster: NM_079175; Mamestra brassicae: AB251894; Chilo suppressalis: AB206477; Tigriopus japonicus: EU831278; Chiromantes haematocheir: AY528900.1; Eriocheir sinensis: EU809924; Portunus trituberculatus: FJ392027; Fenneropenaeus chinensis: EF032650; Penaeus monodon: FJ855436; Metapenaeus ensis: EF470246; Euphausia superba: contig00022. Only the ptHSP90-1 and EusHSP90-1 are used in this analysis.