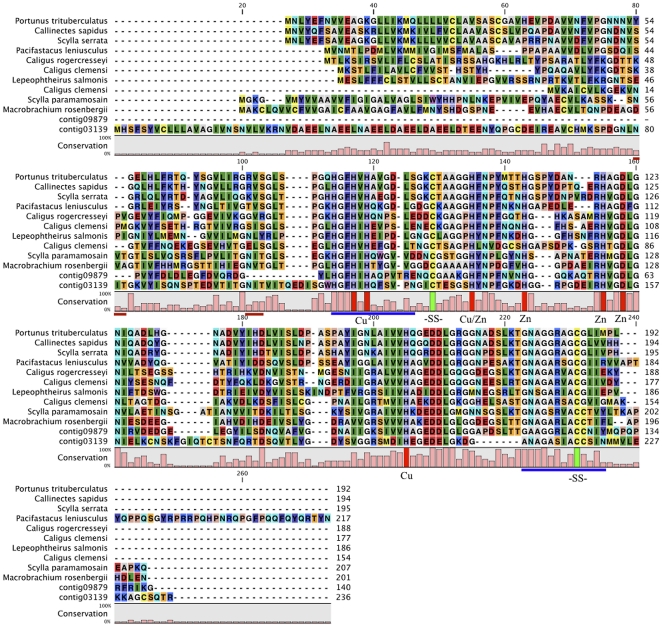
Figure 4. Multiple sequence alignment of the two putative E. Superba Cu-Zn superoxide dismutases with those of other Eucrustacea.
Signature motifs for Cu-Zn superoxide dismutases are indicated by the blue lines under the consensus. The four amino acid residues required for binding copper and four amino acids required for binding zinc are indicated by arrows, as well as two cysteines residues (SS) for the disulphide bridge [110]. Putative N-glycosylation sites for contig03139 only are indicated by the red lines under the consensus and the predicted signal peptide (again for contig03139 only) is marked in italics and underlined. Accession numbers: Portunus trituberculatus: C8XTB0; Callinectes sapidus: Q9NB64; Scylla serrata: A5Y446; Pacifastacus leniusculus: Q9XYS0; Caligus rogercresseyi: C1BQG7; Caligus clemensi: 1: C1C108, 2: C1C2E3; Lepeophteirus salmonis: C1BSR9; Scylla paramamosain: D2DSH2; Macrobrachium rosenbergii: Q45Q33.