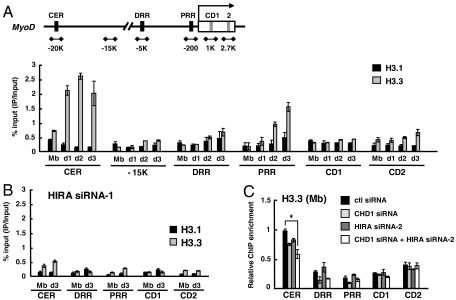
Fig. 3.
H3.3 is incorporated into MyoD regulatory regions in a HIRA-dependent manner. (A and B) ChIP was performed to monitor incorporation of eH3.1 or eH3.3 at MyoD regulatory and coding regions. Chromatin solution prepared at each indicated time point was immunoprecipitated with anti-FLAG and analyzed in duplicate by quantitative real-time PCR to measure the relative enrichment of eH3.1 or eH3.3 at different loci. The data represent the percentage of ChIP (IP/Input). eH3 ChIP signal was normalized to H3 ChIP. (C) eH3.3 levels in the growing myoblasts treated with various siRNAs were analyzed by ChIP and represented as a relative enrichment (the ChIP value obtained from the CER of control siRNA sample was arbitrarily set to 1). For HIRA k.d. (knock down), data with HIRA siRNA-1 and siRNA-2 were shown for B and C, respectively. The sequences of each siRNA are described in the SI Materials and Methods. The k.d. efficiency of two siRNAs is similar as shown in Fig. S4. Error bars represent standard deviation of three independent experiments. *, P < 0.05. eH3, epitope-tagged H3; CER, core enhancer region; DRR, distal regulatory region; PRR, proximal regulatory region; CD, coding region.