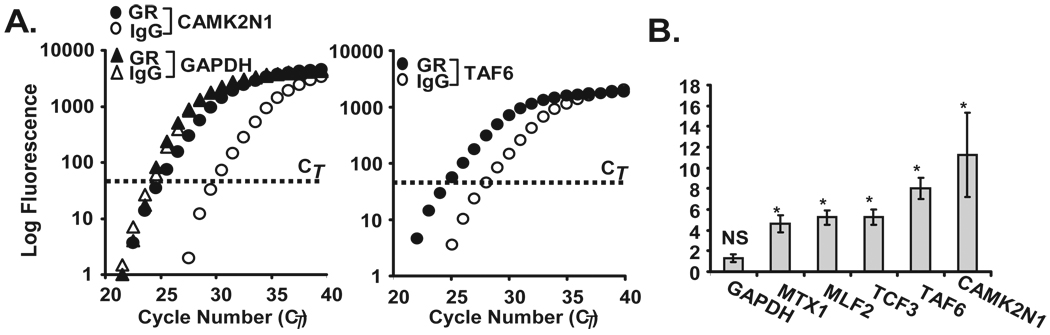
Figure 5. Validation of GR-associated targets identified by gene array.
A. Real-time PCR amplification plot (representative of n=3) after GR (filled circles) or control (open circles) IP for CAMK2N1and TAF6, identified as GR-associated transcripts by the GR-IP array study, and GAPDH as control. B. Bargraph represent the mean ± SEM fold GR/IgG IP enrichment for MTX1, MLF2, TCF3, TAF6, and CAMK2N1 (n=3; *, p<0.05 compared to IgG IP, NS=not significant for GAPDH).