Figure 1. Screening for random mutations.
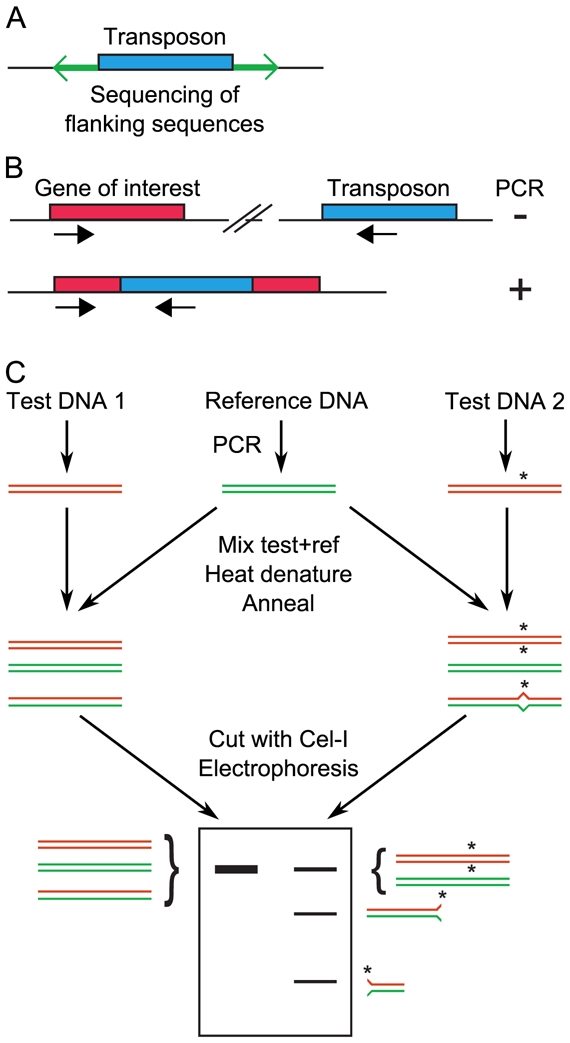
A, Large collections of insertion mutants are generated. For each mutant, the regions flanking the transposon are sequenced, to identify the site of insertion. B, After transposon insertional mutagenesis, screening is achieved by PCR using one primer that hybridizes in the transposon and another that hybridizes in the gene of interest. A PCR product will be obtained only if the transposon is inserted by chance in the gene of interest. To increase throughput, mutants are first tested in pools, then individually for mutants that belong to positive pools. C, TILLING. Regions of interest are simultaneously PCR-amplified from a reference (wild-type) individual and several mutants. Mutant and wild-type amplimeres are mixed together, denatured and reannealed. If a mutation was present in the amplified region of the mutant, then a heteroduplex with one mismatch forms. This heteroduplex is detected by the increased electrophoretic mobility after cleavage by the endonuclease Cel-I.
