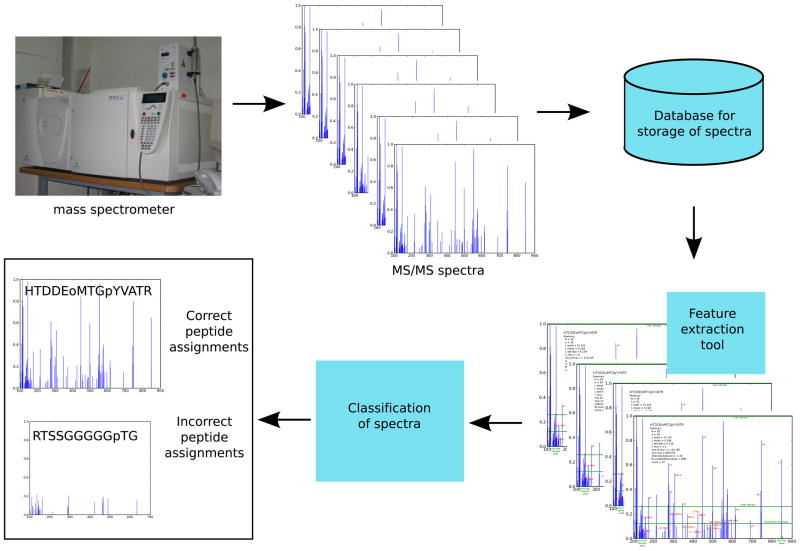
Figure 1.
An overview of phoMSVal. Data are obtained from a phosphospecific mass spectrometry experiment, the spectra are analyzed by Mascot and the data are stored in a database. For each spectrum, features are extracted and used as input into a classifier that separates spectra into two classes: correct or incorrect peptide assignments.