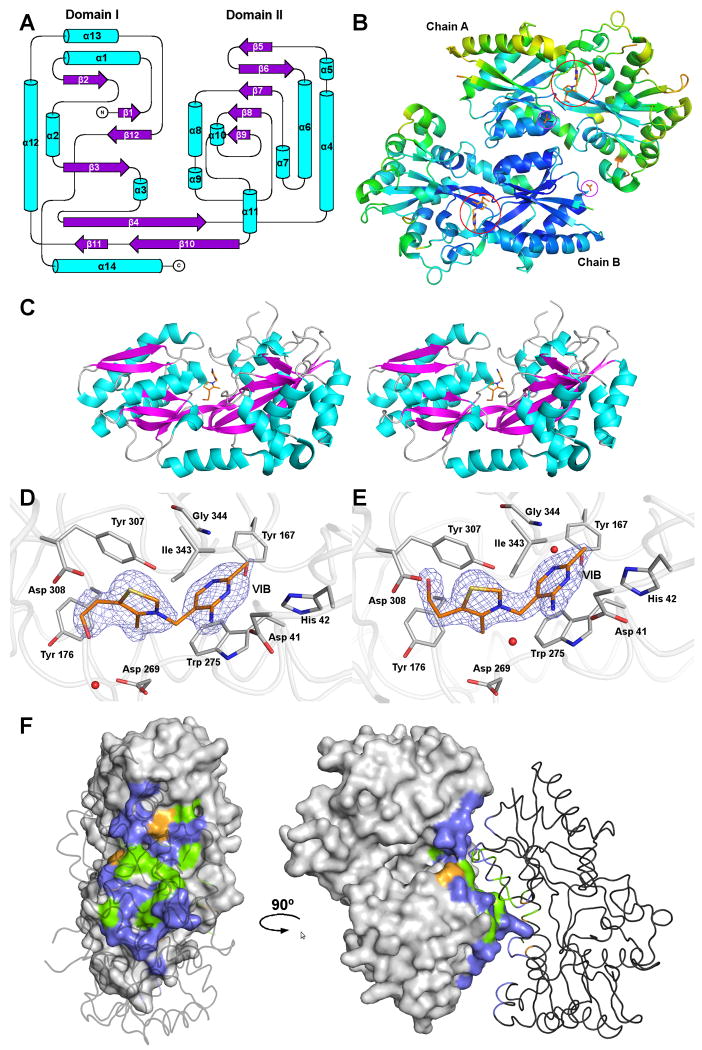
Figure 1.
The structure of MG289.
(A) Secondary structural topology of the MG289 monomer. Helices are colored cyan and strands, magenta. Figure generated in Topdraw 44.
(B) A Cα ribbon trace of the MG289 dimer colored from low to high (blue to red) temperature factor. Ligands are shown as sticks with carbon colored orange; oxygen, red; nitrogen, blue; and sulfur, gold. VIB binding sites are indicated as open red circles. Open purple circles indicate ACT binding sites.
(C) Stereo image of the MG289 monomer with VIB ligand bound. Protein is colored as described in (A), and VIB as described in (B).
(D) The VIB binding site of chain A. The 2Fo-Fc average kick map (mesh) is contoured to 1.0σ 45. Protein carbons are colored grey; ligand carbons, orange; oxygen, red; nitrogen, blue; sulfur, gold. Waters are represented as red spheres.
(E) The VIB binding site of chain B. The 2Fo-Fc average kick map (mesh) is contoured to 1.8σ. Figure colored as described in (D).
(F) The dimer interface. Chain A is shown as a molecular surface, while chain B is a cartoon. Non-interface residues are colored grey; non-polar, green; polar uncharged, violet, and polar charged, orange. All figures generated in PYMOL unless stated otherwise 46.