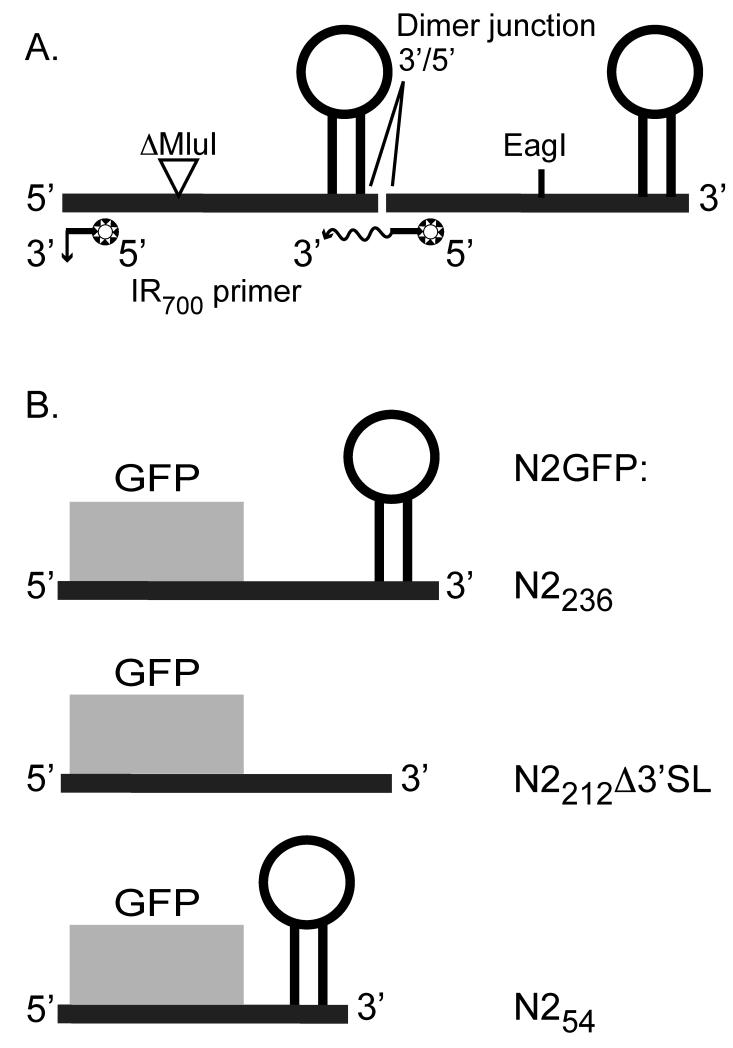
Figure 2. Schematic of RNA transcripts used in these studies.
Panel A: A head-to-tail dimer of NoV RNA2 contains a 319 nt MluI-fragment deletion in the upstream copy and a full-length downstream copy. In vitro transcription is initiated from a T7 promoter in plasmid pT7-N2ΔMluIN2, which is linearized at the unique EagI site shown. The transcript contains two binding sites for the IR dye-labeled primer. On primer binding at the upstream site near the 5′ end, extension of the primer results in a short run-off product, while p31imer binding at the downstream site results in longer extension products that span the dimer junction. Panel B: Replicons N2GFPN2236, N2GFPN2212Δ3′SL, and N2GFPN254 contain 17 nt (nt 1 – 17) from the 5′ end of RNA2 followed by the GFP central core and 236 nt (nt 1100 – 1336), 212 nt (nt 1100 – 1298 and 1323-1336), or 54 nt (nt 1282 – 1336) from the 3′ end of RNA2. Primary transcription of each is initiated in transformed yeast cells from an inducible GAL1 promoter in plasmids pN2GFPN2236, pN2GFPN2212Δ3′SL, and pN2GFPN254, respectively.