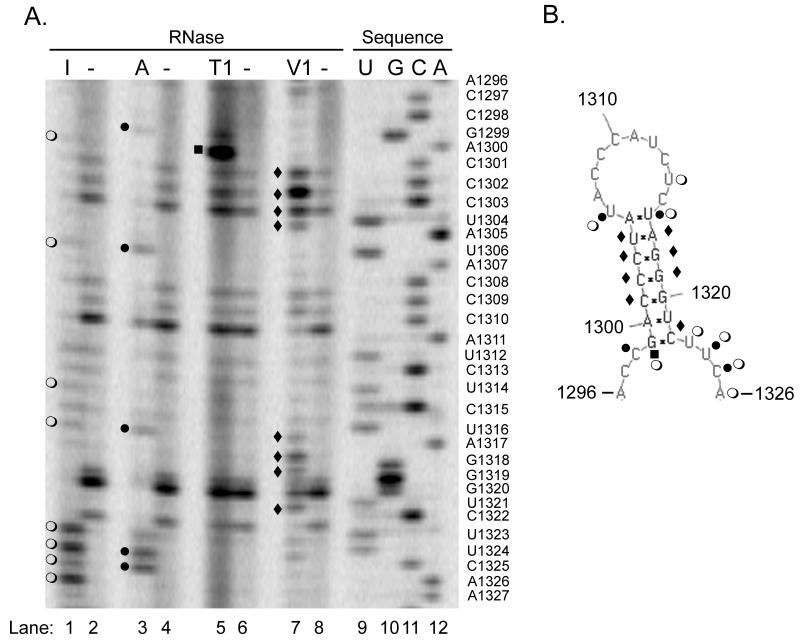
Figure 3. Nuclease mapping studies show the predicted structure can form in solution.
In vitro transcripts of the NoV RNA2 homodimer shown in Figure 2A were synthesized and subjected to digestion with ribonucleases specific for single- (RNases I, A, and T1) or double-stranded RNA (RNase V1), or left untreated. After digestion, samples were subjected to primer extension analysis using an infrared dye-labeled primer complementary to NoV RNA2 nt 57-38. A DNA sequencing ladder was generated with the same primer and the corresponding plasmid DNA template, as described in Material and Methods. Samples were analyzed on a polyacrylamide sequencing gel using a LI-COR 4200 DNA Analyzer. Panel A, nuclease map with sequencing ladder; I, RNase I (lane 1); A, RNase A (lane 3); T1, RNase T1 (lane 5); V1, RNase V1 (lane 7); minus (−), no nuclease (lanes 2, 4, 6, and 8); U, G, C, A (lanes 9 – 12), sequencing ladder. The sequencing ladder was labeled with the nucleotides complementary to the sequence obtained, and U’s were substituted for T’s, to allow direct comparison with panel B. Panel B, predicted structure annotated with the results of the nuclease mapping. Open circles (○), RNase I digestion product; closed circles (●), RNase A; closed squares (■), RNase T1; closed diamonds (◆), RNase V1.