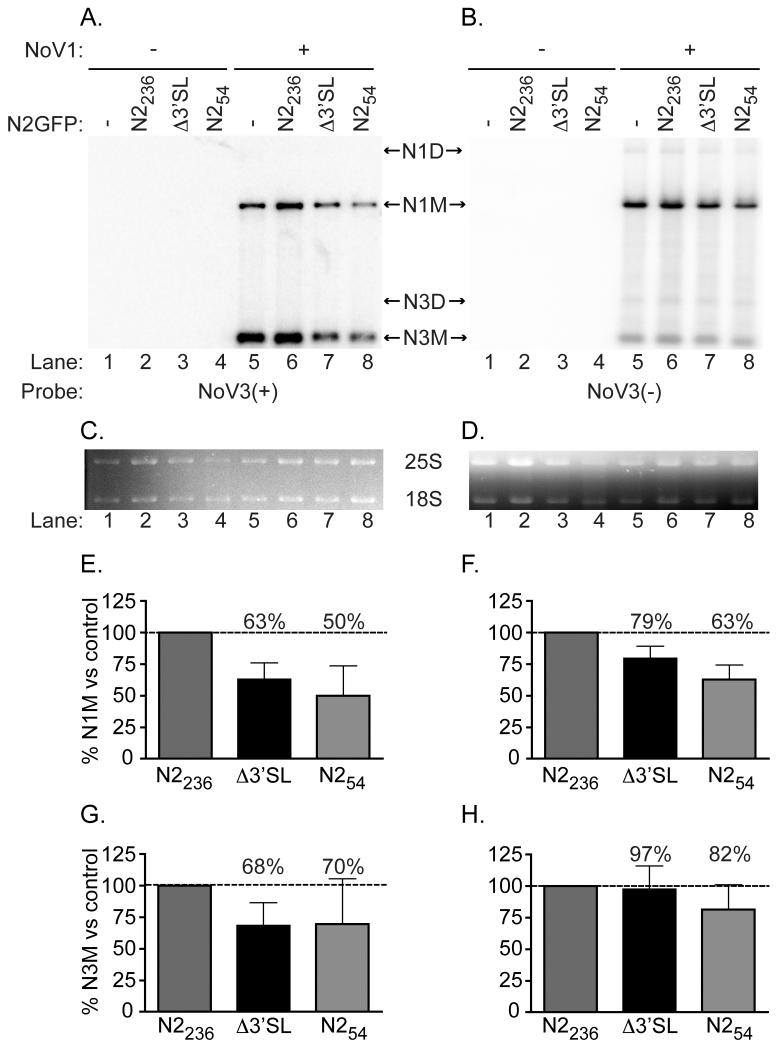
Figure 4. Δ3′SL mutant doesn’t affect RNA1 replication or RNA3 synthesis.
Total yeast RNA isolated from cells transformed with pN2GFPN2236, pN2GFPN2212Δ3′SL, or pN2GFPN254, together with pN1 as a source of RdRp, was separated on a denaturing gel and subjected to Northern blot hybridization analysis using probes specific for the positive (Panel A) or negative (Panel B) strands of RNA3 (labeled as NoV3(+) and NoV3(−), respectively). A representative blot is shown. Prior to transfer, gels were stained with ethidium bromide to allow visualization of yeast cellular 25S and 18S rRNAs (Panels C and D) for use as loading controls. Both monomeric and dimeric forms of NoV RNA1 and RNA3 were detected, indicated in the figure as N1M or N3M (monomers) and N1D or N3D (dimers). Quantitation of the positive- and negative-strand RNA1 and RNA3 species detected, compared to the levels detected for the WT N2GFP236N2 replicon in the presence of RdRp (lane 5) are shown in Panels E–H; : Panel E, RNA1(+); Panel F, RNA1(−); Panel G, RNA3(+); Panel H, RNA3(−). The relative RNA values from three independent experiments are presented as mean values ± standard deviations.