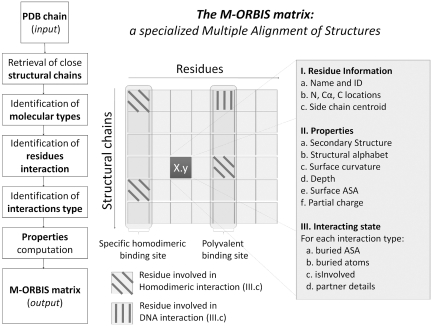
Figure 1.
The M-ORBIS matrix as a specialized multiple alignment of structures. The M-ORBIS output matrix is obtained via a 7-step workflow illustrated in the left part of the figure. As in traditional multiple alignments of sequences or structures, the M-ORBIS matrix gives access to every homologous chain and residue. Nevertheless, to perform the complex task of Molecular Cartography, M-ORBIS also stores information relative to the residue locations (field I.), to the physico-chemical and geometrical properties (field II.) and to the interacting state (field III.) of each analyzed residue. A cell of the M-ORBIS matrix is illustated in the right-hand table and gives insights into some of the properties that are stored. For instance, field III.c indicates that the first residue is involved in only homodimeric interaction in two of six related structural chains. A polyvalent binding site is detected when for a given residue (column), the corresponding residues of related structural chains are involved in at least two different binding site types (residue described in column 5). A specific binding site is inferred when only one binding site type is detected at that position (residue described in column 1).