Figure 1.
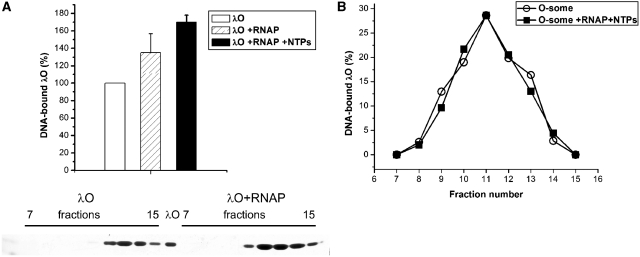
RNA polymerase and transcription process enhance λO binding to oriλ. (A) Formation of O-some in the presence or absence of RNA polymerase activity was analyzed by gel filtration. Nucleoprotein complexes were formed in the presence of supercoiled λ plasmid DNA (10 µg) and λO protein (1.5 µM) and RNA polymerase (2.5 µg) and nucleotides (2 mM each) (as indicated). When the influence of either RNA polymerase or transcription process on O-some formation was assessed, RNA polymerase was first incubated with plasmid DNA and nucleotides (as indicated) for 10 min at 30°C, and then the λO protein was added and incubation was prolonged for another 10 min. Samples were loaded on a Sepharose 4B column; fractions were collected and subjected to SDS–PAGE, followed by silver staining and densitometry. The panel below the graph shows an example of SDS–PAGE separation of fractions obtained after gel filtration of the λO–DNA complexes formed in the presence or absence of RNA polymerase (as indicated). (B) Stability of λO–DNA complex, formed and isolated as described above, was assessed in the presence and absence of transcription. Fractions containing O-some obtained after first round of gel filtration were pooled and divided into half. One of the samples was supplemented with RNA polymerase (2.5 µg) and NTPs (2 mM). Both samples were subsequently (separately) subjected to a second round of gel filtration. Fractions containing O-some were collected and analyzed by SDS-PAGE as described earlier. The quantity of λO protein present in the sample where RNA polymerase and nucleotides were absent was assumed as 100%, and elution profile of both samples was presented according to this value.