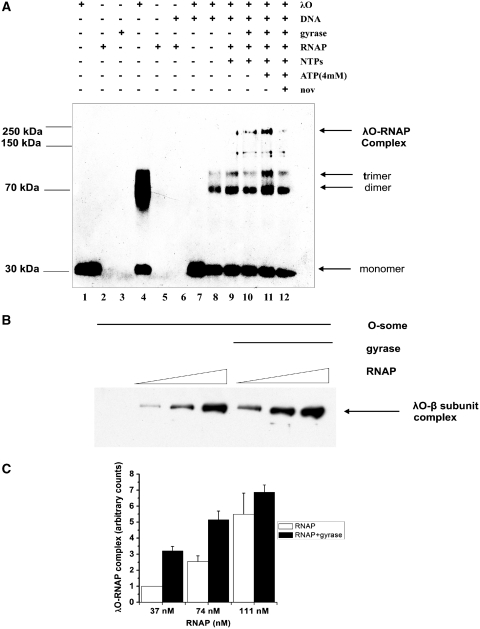
Figure 5.
Gyrase activity stimulates the formation of a complex between RNA polymerase and the λO protein in an ATP-dependent manner. (A) In the presence of λO (0.5 µM) and the supercoiled λ plasmid DNA (2.8 nM) O-some structure was formed and separated from unbound protein fraction by gel filtration. Next, one of the samples was supplemented with additional ATP (final concentration of 4 mM) and novobiocin was added to the reaction mixture to a final concentration of 100 nM. Subsequently, E. coli RNA polymerase (22 nM), gyrase (9 nM) and NTPs (0.5 mM) (as indicated) were added and after a 10-min incubation at 30°C protein complexes were cross-linked with DSG. Complexes were detected by SDS–PAGE followed by immunoblotting with antibodies against λO. λO–RNAP polymerase complex was depicted by an arrow as well as oligomeric forms of the λO protein. The presence or absence of each reaction component was indicated. Lanes 1–3 and 6 contain uncross-linked λO, RNAP and gyrase and O-some, respectively. (B) Experiment was performed analogously as in point A, but higher concentration of the cross-linking reagent was added (1 µM) and various ratios of RNAP to λO were used, as indicated, in the presence of constant concentration of gyrase (8.3 nM), NTPs (500 µM) and ATP (4 mM). (C) λO–RNAP polymerase complex formation in the presence of gyrase activity was measured as described in point B in three independent experiments. Relative amount of λO–β subunit complex formed in the presence and absence of gyrase activity was measured densitometricaly and presented as a function of RNA polymerase concentration with error bars representing SD. Amount of the complex observed in the sample where the lowest concentration of RNA polymerase was present and no gyrase was added was assumed as 1 and other values reflect this value.