Abstract
Zinc finger nucleases (ZFNs) facilitate tailor-made genomic modifications in vivo through the creation of targeted double-stranded breaks. They have been employed to modify the genomes of plants and animals, and cell-based therapies utilizing ZFNs are undergoing clinical trials. However, many ZFNs display dose-dependent toxicity presumably due to the generation of undesired double-stranded breaks at off-target sites. To evaluate the parameters influencing the functional specificity of ZFNs, we compared the in vivo activity of ZFN variants targeting the zebrafish kdrl locus, which display both high on-target activity and dose-dependent toxicity. We evaluated their functional specificity by assessing lesion frequency at 141 potential off-target sites using Illumina sequencing. Only a minority of these off-target sites accumulated lesions, where the thermodynamics of zinc finger–DNA recognition appear to be a defining feature of active sites. Surprisingly, we observed that both the specificity of the incorporated zinc fingers and the choice of the engineered nuclease domain could independently influence the fidelity of these ZFNs. The results of this study have implications for the assessment of likely off-target sites within a genome and point to both zinc finger-dependent and -independent characteristics that can be tailored to create ZFNs with greater precision.
INTRODUCTION
Zinc Finger Nucleases (ZFNs) are artificial restriction enzymes that hold tremendous potential for the manipulation of genomes in a wide variety of plants and animals (1). These enzymes generate a site-specific double-stranded break (DSB) that can abrogate gene function through imprecise repair (via generation of a frameshift) or can introduce tailor-made changes by stimulating homology directed repair from an exogenously supplied DNA template. The utility of ZFNs for gene inactivation and genome editing has been demonstrated in a wide variety of cell lines (2,3), including human ES cells and iPS cells (4,5), as well as in the germline of plants (6–9) and animals (10–15). Due to their demonstrated utility, ZFN-based therapies are being evaluated in clinical trials (16,17).
ZFNs are composed of two modular domains: a tandem array of Cys2His2 zinc fingers (ZFP) tethered to the cleavage domain of FokI endonuclease (Figure 1) (18). The incorporated ZFPs can be engineered to recognize a specific DNA sequence (11,19–21), thereby targeting the attached nuclease domain to a desired location within the genome. Dimerization of the cleavage domain is required for enzymatic activity (22). As a consequence, a pair of ZFNs must bind with the proper orientation and spacing to generate a DSB (23,24). ZFN-mediated gene inactivation/modification is sufficiently robust to generate cell lines with multiple biallelic knockouts (25) and, when applied directly in vivo, founder animals that transmit mutant alleles to their offspring with high frequency (10–15). However, in many instances cytotoxicity is observed as a side effect of ZFN treatment, which presumably results from ZFN-generated DSBs at off-target sites within the genome (11,26–28).
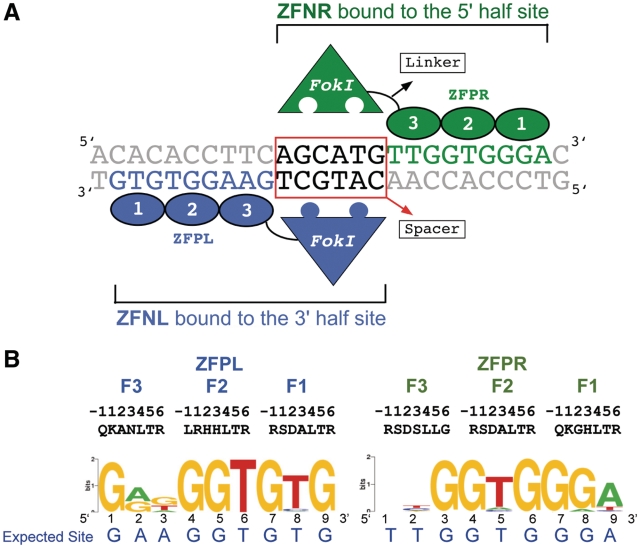
Figure 1.
Zinc finger nucleases targeting exon2 of kdrl: (A) A schematic drawing showing the ZFNs bound to the target site. The two ZFN monomers (ZFNL and ZFNR) bind respectively to the 9 bp 5′- and 3′-half sites through the associated ZFPs (fingers indicated by numbered ovals), which position the heterodimeric nuclease domain over the 6 bp spacer between the two ZFP half sites. (B) The DNA-binding specificities of the kdrl ZFPs determined at high stringency (5 mM 3-AT) using the B1H system displayed as a sequence logo (46,47).
Efforts to improve the in vivo precision of ZFNs have focused primarily on properties influencing DNA recognition. For each ZFN, the number of binding sites within a genome is primarily dictated by the number and quality of the incorporated zinc fingers. Consequently, utilizing ZFPs with higher specificity can reduce the cytotoxicity of ZFNs (28). The type of nuclease domain dictates the active ZFN configurations. ZFNs bearing engineered nuclease variants that preferentially heterodimerize display reduced toxicity in vivo by disfavoring homodimeric DNA recognition (29,30). The number of functional target sites is also defined by the composition and length of the linker joining the ZFP and nuclease domain, which determines the required spacing between ZFN half-sites for activity (23,24). Finally, restricting the in vivo half-life of ZFNs can also attenuate their cytotoxicity (31).
Although the in vivo precision of ZFNs has been analyzed via the characterization of off-target lesion events, an in depth analysis of ZFN properties that influence these effects has not been performed. Potential off-target sites are typically defined by using the DNA-binding specificity of the incorporated ZFPs to scan the genome for sites most similar to these recognition sequences with the appropriate spacing for nuclease activity (5,11,13,16). In the majority of these studies, ZFN-induced lesions are identified at these off-target loci by Cel 1 nuclease or restriction fragment length polymorphism (RFLP) assays (5,12,13). Most of these studies did not detect lesions at their predicted off-target sites, however they typically examined only a small number of off-target sites (<10). Moreover, these assays are not sensitive enough to detect lesion frequencies at ≤1% (16). In two studies (11,16), massively parallel sequencing technology has been used to characterize ZFN-induced off-target lesions with greater sensitivity. Both of these studies revealed that, although infrequent, lesions were present at a subset of the analyzed sites. However, only a small number of off-target sites were analyzed between these two studies: seven heterodimeric sites for the CCR5 ZFNs and 17 for the kdrl ZFNs. Moreover, the influence of ZFN properties on in vivo precision was not examined in either of these studies.
The kdrl ZFNs, which display a low but measurable frequency of off-target events (11), provide an excellent system for exploring the parameters that affect ZFN precision in vivo. In our present study, we performed an in-depth analysis of ZFN precision by assaying lesion frequencies at 141 potential off-target sites in the zebrafish genome. The kdrl ZFNs generate lesions at a small subset of these sites and demonstrate greater promiscuity with increasing dose. Unexpectedly, we found that both the ZFP specificity and dimerization interface of the nuclease domain can influence the precision and activity of ZFNs. These results provide a broader picture of factors that influence the precision of ZFNs with implications for the best compositions to employ for genome manipulations in both model organisms and clinical gene therapy.
METHODS
Zebrafish husbandry
Zebrafish adults and embryos were handled according to standard methods (32). These studies were approved by the UMass Medical School IACUC. The wild-type line used in this study (referred to as Crawfish) was established through several incross generations of wild-type fish originally obtained from Scientific Hatcheries.
ZFN mRNA injections and on-target lesion analysis
ZFPs were cloned into the pCS2 vector containing either DD/RR [R487D (DD) and D483R (RR)] (29,30) or EL/KK [Q486E; I499L (EL) and E490K; I538K (KK)] (29) variants of FokI nuclease as described (11). pCS2-ZFN constructs were linearized with NotI enzyme and mRNAs were transcribed using the mMessage mMachine SP6 kit (Ambion) followed by DNAse treatment. ZFN mRNAs were injected into one-cell stage zebrafish embryos according to standard methods (33). ZFN-induced on-target lesions at the kdrl locus were detected by NspI digestion as described previously (11).
Identifying ZFPs with improved specificity
The DNA-binding specificity of additional clones obtained from B1H-selections for ZFPL in Meng et al. (11) were previously characterized using the 28 bp randomized library via omega-based B1H-selections. The improved ZFPR clone was generated by design incorporating specificity determinants into finger 3 that would be compatible with the desired DNA-binding specificity (34) and its binding specificity was characterized with the B1H system using the 28 bp randomized library. Binding sites from a few surviving clones were sequenced and motifs were generated using MEME (35). Clones (nZFPs) for ZFPL and ZFPR showing improved specificity over the original ZFP (oZFPs) were used for further analysis.
Selection of off-target sites stage I
The zebrafish genome (Zv7 repeat masked) was scanned using a perl algorithm to identify off-target sites containing half-sites similar in sequence to the determined binding site specificities of the two ZFPs (oZFPL: GANGGTGTG and oZFPR: NNGGTGGGA, where N allows all bases) in proper orientation with either 5- or 6-bp spacing between the two half-sites. The sites were ranked based on the number of matches to the target site with a score of 15 matches being the maximum. Heterodimer off-target sites that match the target site at 14 of 15 positions were chosen for analysis (kdrl exon 2 is the only 15 of 15 bp match). Homodimeric sites were derived from sites that match either the ‘GANGGTGTG’ composite site at 14 or 15 out of 16 bp or the ‘NNGGTGGGA’ composite site at 14 of 14 bp. For the pilot scale analysis, a total of 20 heterodimeric and 28 homodimeric sites were chosen. The details for these sites are provided in Supplementary Table S1 online and are marked as ‘off-target sites I’.
Selection of off-target sites stage II
Computational analysis was performed to bin additional off-target sequences (identified as described above) based on the number of conserved guanines (maximum = 10) within the potential off-target sites. A total of 47 sites that contain all 10 guanines were chosen for analysis with a range of total base matches to the target site (13–16 for 5- and 6-bp spacing sites and 14–17 for 14-, 15- and 16-bp spacing sites). Another 47 sites that are missing one or more guanines were chosen for analysis with a range of total base matches to the target site (13–16 for 5- and 6-bp spacing sites and 14–17 for 14-, 15- and 16-bp spacing sites). Both groups are designated in the Supplementary Table S1 (‘off-target sites II—10g’ and ‘off-target sites II—non-10g’). One off-target site is identical between the sites analyzed in Stages I and II.
Solexa data analysis for off-target site lesions
Sequence reads (36-bp) from each Illumina run (both Stages I and II) were binned to different ZFN treatments based on the barcode sequence. For each ZFN treatment, sequences for different off-target sites and the target site were classified using a unique 9 bp ‘prefix’ following the adapter sequence (Supplementary Table S1).
For each off-target site, insertions or deletions in the spacer region were defined based on the distance between the 9 bp ‘prefix’ at the 5′-end of each off-target site and a 6 bp (8 bp in one case) ‘suffix’ at the 3′-end of each off-target site, where a more proximal suffix was employed to identify insertions and a more distal suffix for deletions. In some cases, single nucleotide polymorphisms were present within the suffix sequences requiring a more relaxed suffix sequence definition. If the distance between the prefix and any suffix pair in each sequence matched the expected distance these sequences were binned as ‘correct (W)’, where a secondary distal suffix was also employed to identify sequences of the appropriate length. Distances that were greater than expected were binned as ‘insertions (I)’, and distances that were shorter were binned as ‘deletions (D)’ with the exception that 1 bp insertions or deletions were ignored because of the noise in the sequencing data associated with 1 bp frameshifts in sequences evident in uninjected samples. Reads that did not contain the suffix sequence were marked as undefined (U). This analysis will miss long insertions or deletions that alter either the prefix or suffix but it is robust to the bulk of sequencing errors yielding high-confidence indels. The number of sequencing reads that are correct and the number of reads containing indels (insertions plus deletions) at each analyzed site for each ZFN dose were computed for the subsequent statistical analysis.
All statistical analyses were performed using R, a system for statistical computation and graphics (36). The lesion frequency and its 95% confidence interval (CI) for each off-target site and the target site within each treatment were estimated based on a binomial distribution. The Fisher's exact test was applied to assess whether there is a significant difference between each individual ZFN treatment and the uninjected control in the lesion frequency rate for the on- and off-target sites. The odds ratio and its 95% CI were computed for each ZFN dose using the Fisher's exact test based on conditional maximum likelihood estimation. To adjust for multiple comparisons, P-values were adjusted using the Benjamini–Hochberg (BH) method (37).
Criteria for defining an active off-target site
An off-target site was considered active only if the following criteria were fulfilled: (i) indels occurred at a significant frequency in the injected sample relative to the uninjected control (BH adjusted P-value < 0.05) (37); (ii) indels constituted ≥0.1% of the sequence reads in the average of the two replicates (when applicable); and (iii) more than one different indel sequence was observed (to avoid potential jackpot effects).
Comparing the reproducibility between the two biological replicates of oZFN treatments
To examine the reproducibility of the data, the Pearson correlation test was applied to common sequences between the replicate data sets (oZFN DD/RR replicate 1 and 2: 10 pg normal, 10 and 20 pg deformed) on the log odds ratio of ZFN-treated sample versus control sample (Supplementary Figure S3). The indel rates for the two replicates were averaged for further analyses. If for any off-target site, there were less than 1000 sequences in one of the replicate, the frequency of lesions from the other replicate was used for analysis.
Bacterial one-hybrid based activity assay
To assay the importance of Guanine contacts for the binding of oZFPL and oZFPR to their respective binding sites, a B1H activity assay was performed. The ZFP binding sites (wild-type or mutant) were cloned in the pH3U3 reporter vector. The oZFPL and oZFPR were cotransformed with plasmids bearing their binding sites in US0 cells and the activity assay was performed as described in Noyes et al. (38). From a stock of 109 cells/ml, the 10-fold serial dilutions were placed as 5 µl drops on 2xYT or NM selective media plates containing kanamycin (25 µg/ml), carbenicillin (100 µg/ml), 3-aminotriazole (1 or 10 mM) and IPTG (10 µM). The colonies were grown at 37°C for 20 h (1 mM 3-AT plates) or 48 h (10 mM 3-AT plates). The number of colonies was counted for the NM selective plates and reduction in colony counts was calculated as –log (number of colonies for the wild-type or mutant binding site/ number of colonies for the wild-type sequence).
Additional experimental information is located in the Supplementary Data.
RESULTS
Off-Target analysis of Meng et al. kdrl ZFNs
In our previous study, we demonstrated the efficacy of ZFNs targeting the kdrl gene in zebrafish, which incorporated ZFPs optimized through bacterial one-hybrid (B1H) selections and the ‘DD/RR’ engineered heterodimeric nuclease domain (11). As an initial assessment of the off-target lesions produced by these nucleases, we assayed the presence of lesions at 41 off-target sites (17 heterodimeric and 24 homodimeric), which revealed four heterodimeric off-target sites that accumulated lesions at a low frequency (∼1%). However, the small number of off-target sites examined and the small number of sequences analyzed per site (approximately 250) provided only a limited overview of the off-target activity in the genome.
In order to assess ZFN off-target activity in greater depth, we determined lesion frequencies generated by the kdrl ZFNs at the target site and off-target sites. The off-target sites were chosen based on the DNA-binding specificities of the ZFPs (ZFPL and ZFPR), which we previously determined using the B1H system (11). To provide greater complexity to these motifs, we repeated B1H selections and sequenced the binding sites from the pool of surviving colonies by Illumina sequencing, where more than 1000 unique sequences were used to generate the binding site logo for each ZFP (Figure 1B). Based on these more informative motifs, we found that Position 3 in the ZFPL motif and Positions 1 and 2 in the ZFPR motif provide limited discrimination in DNA recognition. Consequently, these positions were not considered when identifying the most favorable potential off-target sites within the genome based on matches to the target sequence. Based on the ZFPL and ZFPR binding specificity, we chose to characterize 141 putative off-target sites in the zebrafish genome that contained from one to five mismatches relative to the target site (Figure 2A and Supplementary Table S1). Twenty-eight of these sites represent potential recognition sequences for homodimeric ZFNs to examine the exclusivity of the engineered DD/RR nuclease domains. Among the remaining 113 heterodimeric sites, 59 contained the conventional 5 or 6 bp spacer between the two ZFN half-sites. The remaining 54 heterodimeric off-target sites contain a 14, 15 or 16 bp spacer, as previous studies have indicated linker-dependent ZFN activity at sites with longer gaps between the half-sites (23,24).
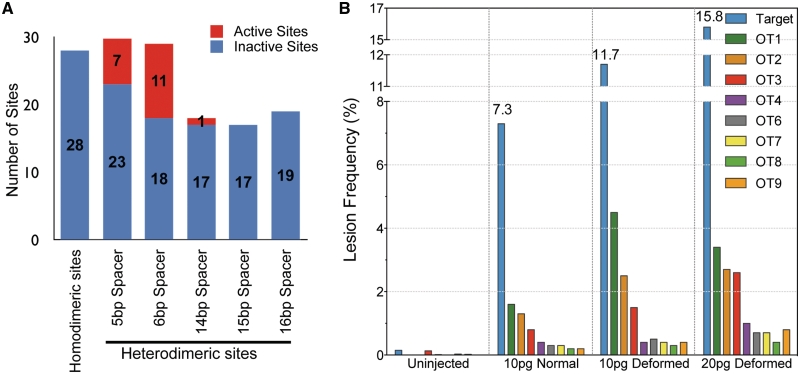
Figure 2.
Overview of the off-target analysis for the original kdrl ZFNs. (A) The number of active (red) and inactive (blue) off-target sites is depicted in the graph. The sites are subdivided according to the type of site (homodimeric or heterodimeric), where heterodimeric sites were divided into five different groups based on the spacing between the two half-sites. A total of eight active off-target sites (see text for criteria) were found in normal embryos from 10 pg ZFN dose (Table 1), and 11 additional off-target sites were active either only in deformed embryos from 10 or 20 pg ZFN dose or in one of the two biological replicates, as described in the text. (B) Dose-dependent effects of kdrl-ZFNs on its in vivo activity and precision. The lesion frequency was plotted for the on-target (blue) and eight off-target sites active in the morphologically normal embryos at the 10 pg dose.
To assess activity of ZFNs at these sites, zebrafish embryos were injected with two different doses of mRNAs (10 or 20 pg) encoding the kdrl ZFNs. These embryos were scored for viability and morphology at 24 h post-fertilization (hpf) to provide an overt assessment of toxicity (Supplementary Figure S1). At the 10 pg dose, ∼50% of the surviving embryos were morphologically normal whereas the remainder displayed developmental abnormalities (‘deformed’ henceforth). Separate pools of ∼25 injected embryos were prepared from morphologically normal and deformed embryos for lesion analysis. At the 20 pg dose, the majority of embryos were deformed or dead. Consequently, only deformed embryos were characterized at this dose. RFLP analysis confirmed the activity of kdrl ZFNs at the target site (Supplementary Figure S2).
The presence and frequency of lesions at each site was determined by illumina-based sequencing of PCR amplicons spanning each genomic locus. On average, approximately 8000 reads per site were obtained, which allowed confident assessment of combined insertion and deletion (indel) frequencies ≥0.1%. Owing to the short-read length of illumina sequencing (∼36 bp), our analysis was limited to the detection of small insertions or deletions. Only indels that were >1 bp in length were counted to avoid the bulk of the sequencing artifacts. To ascertain the consistency of the data, lesions at a subset of off-target sites were analyzed from a second independent biological replicate of the ZFN injections. Analysis of the site-specific lesion frequency between the biological replicates shows that they are significantly correlated (Supplementary Figure S3). The presence of indels at each site was considered significant only if the following criteria were fulfilled: (i) indels occurred at a significantly higher frequency (BH adjusted P-value < 0.05) in the injected sample relative to the uninjected control to account for noise in the sequencing data at some sites, which leads to a small fraction of sequences that appear to contain lesions even in the uninjected control (37); (ii) indels constituted ≥0.1% of the sequence reads (in the average of the two replicates where available); and (iii) more than one different indel sequence was observed (to avoid potential jackpot effects). We believe these criteria constitute a conservative assessment of activity, and may assign sites as inactive that actually incur indels at a low frequency. Consistent with the RFLP analysis of the kdrl ZFNs, the lesion frequency at the target site was ∼7% in normal embryos at the 10 pg dose, which increased to ∼15% at the 20 pg dose (Table 1).
Table 1.
Sequences and lesion frequencies for each ZFN pair and dose at the target and 19 active off-target sites
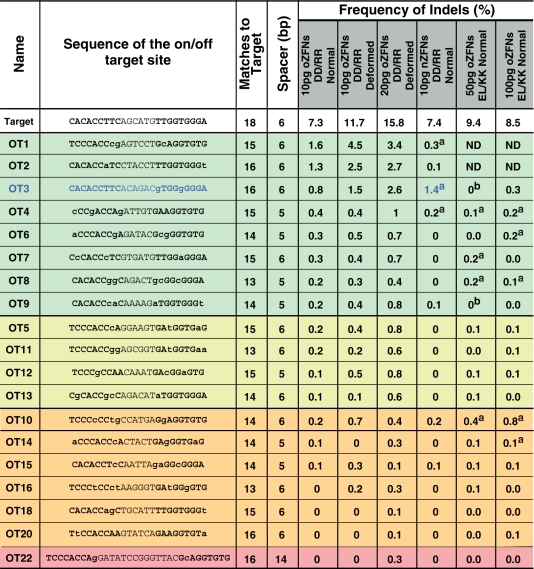 |
Off-target sites show significant lesion frequencies either for normal embryos (green) or only in deformed embryos (yellow) injected with original ZFNsDDRR (oZFNsDDRR). Off-target sites in orange displayed significant activity in only one of two biological replicates. The red off-target site has a 14 bp spacer separating the ZFP recognition sequences. Bold sequences within the on- and off-target sites indicate putative ZFP binding sites. ND = not determined.
aIndicates off-target sites in the final three columns that meet our significance criteria. Only one off-target site showed increased lesion frequency in nZFNs as compared to oZFNs-DD/RR (OT3 in Blue).
b‘0’ in the 50 pg oZFNELKK column indicates two off-target sites which had insufficient sequencing reads to determine the lesion frequency.
Overall only 19 off-target sites were ‘active’ (i.e. displayed indels at a significant frequency based on the criteria above) even at the higher ZFN dose (Figure 2A). All of the examined homodimeric sites were inactive, which is consistent with previous studies indicating that the DD/RR nuclease domain suppresses activity at homodimeric sites (11,30). In ZFN-treated embryos with normal morphology, only eight of the 113 heterodimeric sites were active (across both biological replicates where available, Figure 2B and Table 1). Notably, all of these sites contain a 5 or 6 bp spacer between the two half-sites. At the higher ZFN dose, an additional four off-target sites were actively cleaved. Moreover, seven other off-target sites were found to be active in one of the two biological replicates. Since these seven sites contained hallmarks of ZFN-induced lesions (multiple types of lesions in the spacer region between the two ZFN half-sites), we included them in our analysis of active sites. One of these sites (OT22 in Table 1) contains a longer spacer (14 bp) between the ZFN binding sites. Among the examined off-target sites, those containing a 6 bp spacer were the most likely to be active, both based on the fraction of active sites (38%) and the indel rates at active sequences (Figure 2A and Table 1). Off-target sites containing a 5 bp spacer were the only other group where multiple sites (23%) were actively cleaved. These results are consistent with a previous study indicating that ZFNs with a ‘TGGS’ linker connecting the ZFP and the nuclease domain are most active on target sites separated by a 6 bp spacing followed by sites with a 5 bp spacing, whereas sites with longer spacers are inefficiently cleaved (24). With regards to the types of observed lesions 4 bp insertions, which represent a simple fill-in and religation of the 5′-overhangs generated by the FokI nuclease domain, are the most common events (Supplementary Figure S4A and B).
Concomitant with greater on-target lesion frequency, increasing the ZFN dose increased the degree of off-target cleavage within the genome (Figure 2B). However, preferential activity at the target site was maintained at both ZFN doses, as the on-target lesion frequency exceeded that of any off-target site by at least 4-fold. Notably, animals treated with the higher ZFN dose were more likely to be deformed suggesting that increased collateral damage within the genome may contribute to their abnormal development. Consistent with this hypothesis, normal embryos at the 10 pg dose displayed fewer active off-target sites than the deformed embryos at the 20 pg dose (8 versus 12 considering active sites in both the replicates or 12 versus 18 considering activity in one replicate). Moreover, these normal embryos also exhibited significantly lower frequencies of off-target lesions at the eight common active off-target sites with the median lesion frequency increasing from 0.6% in normal embryos to 1.5% in deformed embryos at the 20 pg dose (P < 0.0001). Thus, increased off-target lesion frequency is associated with the presence of developmental abnormalities.
Common features of active off-target sites
We next sought to identify common characteristics of active off-target sites that distinguish them from inactive sites. Since active sites could simply share greater homology to the kdrl target sequence, we compared the total number of matches to the target site for active versus inactive off-target sequences containing a 5- or 6-bp gap between the ZFN half-sites (Figure 3A). Surprisingly, there was no significant correlation between the degree of identity and off-target activity (P = 0.48). Furthermore, the distribution of active sites with regard to homology to the target site simply reflected the general distribution of all prospective off-target sites (τ = 0.89, P = 0.0367). Thus, in this population of sites that are highly similar to the kdrl ZFN target sequence, the degree of identity is not a defining feature of activity.
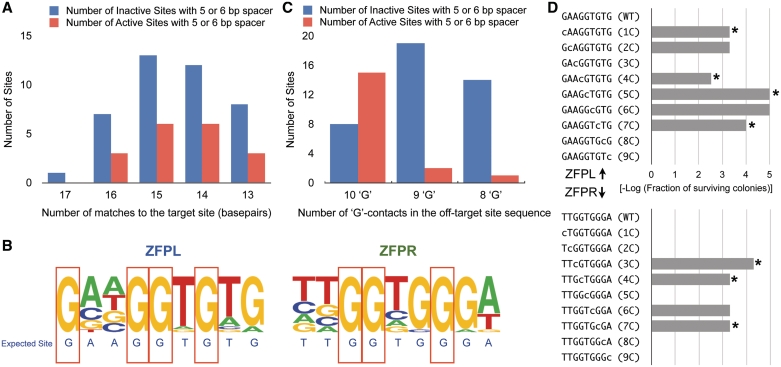
Figure 3.
Characteristics of active off-target sites. (A) The distribution of the number of matches to the target site for active (red) and inactive (blue) off-target sites with 5- or 6-bp spacing is shown. (B) Base frequency at each position in the ZFPL and ZFPR binding sites are displayed as a logo for the group of 18-active off-target sites. Guanines at seven positions (red boxes) in the binding sites were absolutely conserved within these sequences, while they are more variable in the inactive sites (Supplementary Figure S5). (C) The distribution of the number of active (red) and inactive (blue) off-target sites as a function of the number of guanines preserved in each recognition sequence. (D) Each base in the binding site of the ZFPL and ZFPR was independently mutated to cytosine, which is not found at any position in either of the ZFP recognition sequences, and its influence on ZFP binding was assayed using B1H-based activity assay (38) at 1 mM 3-AT. This assay will detect only the most important positions for recognition, where a reduction in cell survival (plotted as the –log of surviving colonies) indicates a position important for recognition. All of the absolutely conserved guanines—indicated by an asterisk—are critical for activity.
To better identify attributes that distinguish active from inactive off-target sequences, we constructed a frequency plot of the bases at each position in the sites from the active group (Figure 3B). One striking characteristic of the active off-target sites is the complete conservation of a number of the guanines (7 of 10) in the composite ZFP recognition sequences. These positions are typically more diverse in the inactive sequences (Supplementary Figure S5), suggesting that they represent critical features that define activity. Examining this trend in greater depth, we find that 15 out of 23 off-target sites with all 10 ‘G’ contacts were active, whereas only three out of 36 off-target sites lacking one or more of these ‘G’ contacts were active (Figure 3C). This correlation was highly significant (P-value = 5.6e−6). Using a B1H-based activity assay (38), we directly determined the importance of the ‘G’ contacts in the kdrl-ZFPL and kdrl-ZFPR recognition sequences by mutating each base independently to cytosine and assaying the effect on ZFP-dependent cell growth. Consistent with the in vivo data, we observed that mutation of any of the conserved Gs in the ZFPL or ZFPR binding site strongly reduced ZFP-dependent cell growth even at low stringency (1 mM 3-AT), where only the most important recognition positions should be detected (Figure 3D). Other positions within each ZFP binding site also influence recognition; however, the impact of mutations at some of these positions is only detected at higher stringency within the activity assay (10 mM 3-AT, Supplementary Figure S6). Thus, for these ZFPs the conserved G contacts identified in this analysis appear to be necessary but not sufficient for efficient recognition of their subsites.
Reduced off-target effects when employing ZFNs with improved specificity
Having established a baseline of off-target events with our original kdrl ZFNs, we investigated the influence of the two distinct functional domains within the ZFN (the ZFP and the nuclease domain) on in vivo precision. We focused initially on further optimization of the kdrl ZFPs since improving their DNA-binding specificity would be expected to have the greatest impact on off-target events. Based on the determined specificity of these ZFPs (Figure 1B), each ZFP displayed a strong preference for the desired base pair at approximately seven of nine positions within their target site. However, in both ZFPs two positions within the C-terminal finger (Finger 3) recognition site were relatively poorly specified, which became our focus for improvement. To identify ZFPs with improved specificity, additional clones generated from our original B1H selections were characterized using B1H-based binding site selections followed by sequencing of binding sites from a few surviving clones (11). This yielded an improved clone (nZFPL) for the left recognition site, but no obviously improved clone was identified for right recognition site. Instead, a modestly improved clone (nZFPR) was generated by introducing previously defined specificity determinants that are compatible with T recognition at Positions 3 and 6 of the Finger 3 recognition helix (34). Subsequent efforts to reselect Finger 3 of the ZFPR in a different context yielded an identical finger sequence (RSDALRK) (C. Zhu et al., unpublished data). Comprehensive binding motifs for the new ZFPs (nZFPs) were determined by B1H binding site selections followed by illumina sequencing. More than 1000 unique sequences were used to generate each recognition motif. Comparison of the recognition motifs indicates improvements in the specificity of both nZFPs (Figure 4a). nZFPL displays a dramatic increase in the preference for adenine at positions 2 (rising from 56 to 99%) and 3 (rising from 13 to 36%) within the recovered sequences. Likewise, nZFPR displays a modest increase in the preference for thymine at position 2 (rising from 42 to 67%) within the recovered sequences.
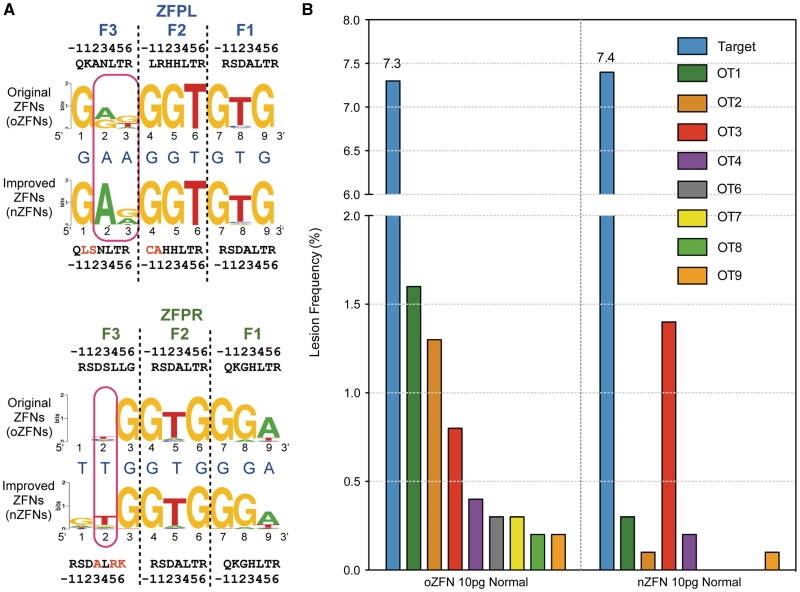
Figure 4.
The specificity of the ZFP domains influences the precision of ZFNs. (A) Binding site specificities of the new and old ZFPs determined using the B1H system displayed as Sequence logos (46,47). The recognition helix sequences for each finger are displayed where the amino acids that differ in the nZFPs are indicated in red. Red rectangles highlight the positions where information content of the desired base was higher in the improved ZFNs. (B) Comparison of lesion frequencies at the on-target site and the 8 active off-target sites were plotted for oZFN- and nZFN-treated embryos. The color scheme remains same as in Figure 2. Lesion frequency at the off-target sites in nZFN-treated embryos was reduced at all but one off-target site.
Although the improvements in specificity of the nZFPs appear modest, we assessed whether these differences would translate into improved ZFN precision in vivo. We compared the in vivo activity and toxicity of the nZFNs (incorporating the new ZFPs) with original ZFNs (oZFNs). mRNAs (either 10 or 20 pg dose) encoding each set of ZFNs were injected into zebrafish embryos. After 24 hpf, treated embryos were scored as morphologically normal or deformed. The nZFNs displayed markedly lower toxicity: ∼45% of the nZFN-treated embryos displayed normal morphology at the 20 pg dose whereas only ∼17% of the oZFNs-treated embryos were normal (Supplementary Figure S1).
We reasoned that the reduced toxicity of the nZFNs was a consequence of decreased off-target cleavage. Therefore, we compared off-target lesion frequencies at the same 141 sites characterized for the oZFNs in genomic DNA isolated from embryos treated with the nZFNs. The nZFNs, at a dose of 10 pg, showed an on-target lesion frequency of ∼7.4%, which was similar to that observed with an analogous dose of the oZFNs. Notably, even with similar on-target activity, nZFNs displayed significantly lower rates (P < 0.0001) of off-target cleavage at the majority (seven out of eight) of the active off-target sites for the oZFNs in normal embryos (Figure 4b and Table 1). Among the 59 heterodimeric off-target sites with a 5 or 6 bp spacer, only three displayed lesions at a significant frequency based on our criteria (Table 1), which represented a reduction compared to the eight active sites for the oZFNs. Only one off-target site (OT3) showed an increase in the lesion frequency with the nZFNs. This may be due to the presence of a 5′-guanine in the nZFPR OT3 half-site, as the nZFP recognition motif indicates a slight preference for ‘G’ at this position in the recognition sequence, which is absent in the oZFPR recognition motif. Thus, based on this analysis even a modest improvement in ZFP specificity can result in dramatic reduction in ZFNs promiscuity.
Examining the influence of the nuclease domain variant on ZFN promiscuity
Although the primary determinant of ZFN specificity is the incorporated ZFP, there is ample evidence that the nuclease domain can also influence the cytotoxicity of ZFNs (29,30). Consequently, the influence of the FokI nuclease dimerization interface on ZFN activity and precision in vivo was investigated. We compared the on- and off-target activity of the original kdrl ZFNs containing the engineered heterodimeric DD/RR nuclease domains (ZFNsDDRR) to the same ZFPs fused to the heterodimeric EL/KK versions of the FokI nuclease domain (ZFNsELKK) (29). Although both nuclease variants have been successfully used on chromosomal targets in vivo (29,30), there has not been a detailed study comparing their activity and their potential influence on ZFN specificity in vivo. Notably, we found that the ZFNsELKK had a markedly lower activity such that injection of 5–10 times more mRNA (50 and 100 pg doses) was required to achieve on-target lesion rates similar to the ZFNsDDRR (Supplementary Figure S2). Consequently we performed lesion analysis for the EL/KK ZFNs at these higher doses, where we examined a subset of the previously characterized off-target sites (96 out of 141). Unexpectedly, we found that the ZFNsELKK displayed reduced off-target lesion frequencies compared with the ZFNsDDRR (Figure 5 and Supplementary Figure S7). At previously defined active off-target sites, normal embryos treated with 100 pg of ZFNsELKK displayed a significantly lower average off-target lesion frequency (0.13%) than normal embryos treated with 10 pg ZFNsDDRR (0.37%, P < 0.0001). Only one other off-target site (OT10, Table 1) consistently displayed significant lesions in the ZFNsELKK-treated embryos. Notably, for this site all 10 of the target site guanines are retained. Thus, for the original kdrl ZFNs, the choice of the engineered nuclease domain has a surprising impact on the ratio of on-target to off-target lesions in vivo.
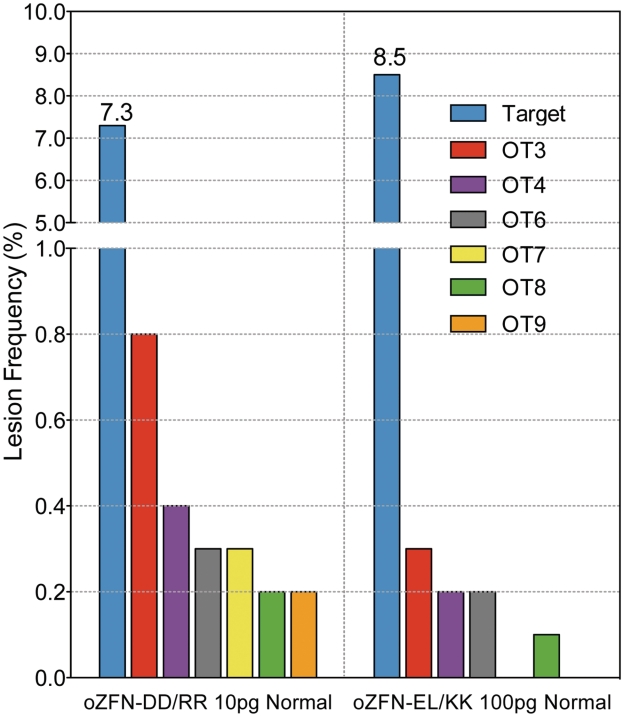
Figure 5.
Influence of the type of the engineered nuclease domain (DD/RR or EL/KK) on the precision of the original ZFNs. Injection of 50–100 pg of mRNA for oZFNELKK was required to achieve on-target lesion frequency similar to 10 pg of oZFNsDDRR. The lesion frequencies for normal embryos treated with 10 pg dose of ZFNsDDRR or 100 pg dose of oZFNELKK were plotted for the on-target site and a subset (six of eight) of the active off-target sites for ZFNsDDRR that were assayed in this experiment.
DISCUSSION
Although ZFNs have been used to create genetically engineered organisms (39,40) and initial clinical trials employing them as therapeutics are underway (16,17,41), the characterization of ZFN-induced collateral damage to the genome of treated cells has been limited primarily to indirect assays of toxicity (26) and DSB foci (16,28) or lesion analysis at a small number of potential off-target sequences (12,13). In this study, we have performed the most detailed analysis to date of the off-target effects of ZFNs by characterizing lesion frequencies at 141 potential off-target sites from the genomes of ZFN-treated zebrafish embryos. Using the kdrl ZFNs as a model, we show that the B1H-selected three-finger ZFNs preferentially cleave their target site to any assayed off-target site and thus, are sufficient for relatively precise gene modification. We also probed the influence of the components of kdrl-ZFNs on their precision. Surprisingly, both the choice of the nuclease domain and the specificity of the component ZFP domains dictate the accuracy of these ZFNs.
Not unexpectedly, the thermodynamics of DNA recognition appear to dominate the impact of binding site mutations on ZFN activity. Simply assessing the likelihood of ZFN activity at an off-target site based on the number of matches to the target sequence was a poor predictor, as evidenced by the absence of correlation within the data for our three-finger ZFNs (Figure 3A). Off-target sites with as many as five mismatches to the target site contained indels at a statistically significant frequency, whereas other sites with just one or two mismatches were inactive. Data from binding site selections provides a much better metric for defining critical positions for recognition. The relative importance of individual positions within each ZFP binding site was initially defined by our high stringency B1H binding site selections (Figure 1B), which provided a consensus recognition sequence for each ZFP. The most critical positions were identified using the low stringency B1H activity assay, where we could examine the importance of individual positions by mutating them independently (Figure 3D). In principle, information on the most critical positions could also be obtained through B1H binding site selections performed at low stringency. In the case of kdrl ZFNs, the preservation of a subset of the arginine–guanine interactions in off-target sites was strongly correlated with ZFN activity at these sequences. Arginine–guanine interactions are typically important specificity determinants at the zinc-finger–DNA interface: abrogating similar contacts in the Zif268 recognition sequence results in a 100- to 400-fold decrease in its binding affinity (42). Based on these observations, we speculate that engineering ZFNs with specificity determinants that distribute the binding energy more uniformly over the entire recognition sequence—instead of employing a few critical arginine–guanine contacts—will result in ZFNs with improved functional specificity. Achieving this goal may require increasing the number of fingers per ZFP as well as the use of appropriate linkers to attenuate ZFP affinity (43), a hallmark of many of the ZFNs currently employed by Sangamo BioSciences (13,16).
The influence of ZFP specificity on the in vivo activity and toxicity of ZFNs was first demonstrated by Cornu et al. (28) where they compared the activities of ZFNs containing modularly assembled ZFPs to ZFNs containing ZFPs selected for the identical target sequences. The selected ZFPs displayed higher specificity as measured by the ratio of the affinity of each ZFP for its target site relative to bulk non-specific DNA. When incorporated into ZFNs, the resulting nucleases generally showed higher activity and lower toxicity in human cells than the nucleases containing their modularly assembled counterparts. In this study, we have performed a more in-depth analysis by defining the base-preferences at each binding site position for the employed ZFPs, which, unlike the bulk specificity, provides information about key sequence features likely to be shared by potentially active off-target sites. This information coupled with a broad assessment of the frequency of ZFN-induced lesions at a number of off-target sites in the genome of zebrafish embryos reveals that even modest changes in the ZFP specificity can decrease off-target activity leading to improved functional specificity and reduced toxicity. Thus, detailed specificity analysis of candidate ZFPs provides not only an estimate of key sequence features of potentially active off-target sites but also an assessment of the relative fitness of the candidate for utilization in ZFNs. In cases where the DNA-binding specificity is sub-optimal, this information can be employed for focused optimization of suspect specificity determinants to obtain ZFPs with higher specificity and superior in vivo performance.
Surprisingly, in addition to the influence of the ZFP specificity on ZFN activity, we observed that the type of the engineered nuclease domain influences ZFN precision. We examined the influence of two pairs of FokI variants DD/RR and EL/KK on ZFN activity, both of which favor heterodimerization over homodimerization (29,30) and display lower in vivo toxicity. Although, Miller et al. reported that ZFNs incorporating these engineered FokI nuclease variants show 2- to 3-fold less activity than the wild-type domain, there has been no detailed study comparing their relative precision. In fact, conflicting data exists regarding the precision of these engineered nucleases. Kim et al. (44) found that only the DD/RR nuclease variant appeared to reduce cellular toxicity relative to the WT nuclease domain. In our study, the kdrl ZFNs harboring the EL/KK variant (ZFNsELKK) consistently show lower activity than the ZFNs harboring the DD/RR nuclease (ZFNsDDRR). Consequently, a 5-fold higher dose of ZFNsELKK was required to obtain an on-target lesion frequency similar to the ZFNsDDRR. This result differs from a recent report by Guo et al. (45) that the EL/KK-containing ZFNs are more active than DD/RR-containing ZFNs on an integrated target in 293 cells. We cannot explain this discrepancy, however, our observation of reduced activity for the EL/KK variants in zebrafish has been confirmed at number of other ZFNs targeting different genomic loci (T. Smith et al., unpublished data). For the kdrl ZFNs, even though EL/KK variants were injected at an elevated dose the toxicity of ZFNsELKK and ZFNsDDRR was similar (Supplementary Figure S1) and genomic analysis confirmed that the ZFNsELKK generate fewer off-target lesions than the ZFNsDDRR. The decreased activity and toxicity of the ZFNsELKK could be the result of lower dimerization potential for the EL/KK nuclease domain, which would reduce the degree of cooperative binding between the two EL/KK monomers. As a result, stronger interactions between each ZFP monomer and its binding site would be required to achieve residence times necessary to generate a DSB. Reduced cooperativity has been previously proposed as an explanation for the decreased toxicity of EL/KK variant as compared to the wild-type nuclease domain (29). However, the reduced toxicity of EL/KK variant in this study could have been associated with its limited homodimeric activity. By directly comparing the EL/KK and DD/RR nuclease variants, neither of which displays significant homodimeric activity based on our analysis, it is readily apparent that the cooperativity between the nuclease monomers is an important feature of ZFN activity. These results suggest that further reduction in the dimerization potential of the nuclease domain coupled with specific zinc fingers with distributed binding affinity may lead to additional improvements in the precision of ZFNs.
ZFNs have been used to create genetically engineered organisms like zebrafish and rats where generating gene modifications with conventional homologous recombination based methods has not been feasible. We and others have shown that these genetic modifications created using ZFNs can be transmitted through the germline. However, the degree of germline transmission of off-target lesions is an unaddressed concern for these ZFN-modified animals. To assess this possibility, we outcrossed one founder fish generated in Meng et al. (11) and examined the progeny for the presence of lesions at the active off-target sites identified in this report. Although, we found lesions at the target kdrl site in ∼50% of 35 offspring analyzed, we did not find evidence of lesions at any of the off-target sites (data not shown). This result, although merely representing a single founder, suggests that using ZFNs generated via B1H-based selections, one can obtain lines of genetically engineered animals relatively free of background mutations without the need for extensive outcrossing of founder animals.
Although this is the most detailed study of the off-target effects of ZFNs to date numerous questions remain to be addressed. One of the key limitations of this study is its characterization of ZFNs specific for a single target sequence. Although this study has improved our understanding of the activity of ZFNs within the genome, further analysis of the activity of other ZFNs pairs will allow a more comprehensive understanding of ZFN activity in vivo. This study is also biased by our choice of genomic sites for analysis based on the characterized specificity of our ZFPs. A more comprehensive survey of active ZFN targets could be obtained by performing a genome-wide analysis of ZFN occupancy using ChIP-seq in combination with lesion analysis, which might identify classes of active target sites (such as alternate spacings or registers of binding) that were uncharacterized in our survey. Ultimately, understanding the parameters that influence the precision of ZFNs in vivo will lead to improved designs facilitating the ease of creating genetically modified organisms as well as improved therapeutics for gene therapy.
ACCESSION NUMBER
Sequencing data from the off-target analysis in vivo is available through GEO (GSE23762).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A.G., L.J.Z., N.D.L. and S.A.W. were supported in part by a grant from the National Heart, Lung, and Blood Institute (1R01HL093766 to S.A.W. and N.D.L.); X.M. and S.A.W. were supported in part by a grant from the National Institute of General Medical Sciences (1R01GM068110 to S.A.W.). Funding for open access charge: National Heart, Lung, and Blood Institute (1R01HL093766).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Ryan Christensen and Gary Stormo for their advice on the computational analysis of ZFP specificity, and to Mike Kacergis for valuable assistance in fish care.
REFERENCES
- 1.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 2.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 3.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nature Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 4.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osakabe K, Osakabe Y, Toki S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2010;107:12034–12039. doi: 10.1073/pnas.1000234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nature Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS ONE. 2010;5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alper J. One-off therapy for HIV. Nature Biotechnol. 2009;27:300. doi: 10.1038/nbt0409-300. [DOI] [PubMed] [Google Scholar]
- 18.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc. Natl Acad. Sci. USA. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat. Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 21.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol. Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu PQ, Chan E, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, et al. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol. Bioeng. 2009;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 26.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 27.Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, 3rd, Segal DJ, Weitzman MD, Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 28.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 29.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 30.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nature Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 31.Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westerfield M. The Zebrafish Book. Eugene, Oregon: University of Oregon Press; 1993. [Google Scholar]
- 33.Xu Q. Microinjection into Zebrafish embryos. In: Guille M, editor. Molecular Methods in Developmental Biology. Vol. 127. Totowa, NJ: Humana Press Inc.; 1999. pp. 125–132. [DOI] [PubMed] [Google Scholar]
- 34.Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 35.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 36.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 38.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Provost F, Lillico S, Passet B, Young R, Whitelaw B, Vilotte JL. Zinc finger nuclease technology heralds a new era in mammalian transgenesis. Trends Biotechnol. 2010;28:134–141. doi: 10.1016/j.tibtech.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Yan Z, Sun X, Engelhardt JF. Progress and prospects: techniques for site-directed mutagenesis in animal models. Gene Ther. 2009;16:581–588. doi: 10.1038/gt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elrod-Erickson M, Pabo CO. Binding studies with mutants of Zif268. Contribution of individual side chains to binding affinity and specificity in the Zif268 zinc finger-DNA complex. J. Biol. Chem. 1999;274:19281–19285. doi: 10.1074/jbc.274.27.19281. [DOI] [PubMed] [Google Scholar]
- 43.Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc. Natl Acad. Sci. USA. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J, Gaj T, Barbas CF., 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J. Mol. Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.