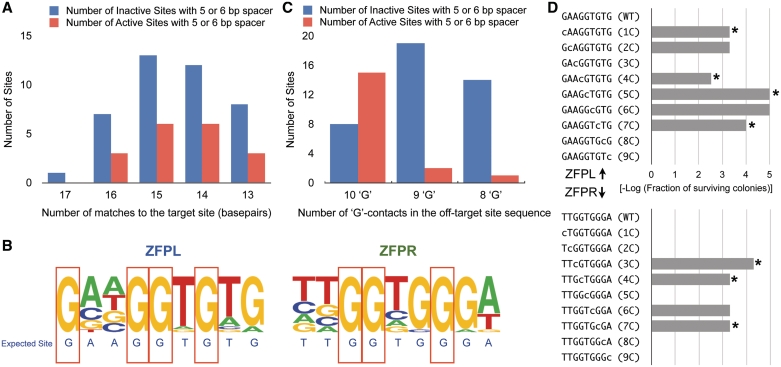
Figure 3.
Characteristics of active off-target sites. (A) The distribution of the number of matches to the target site for active (red) and inactive (blue) off-target sites with 5- or 6-bp spacing is shown. (B) Base frequency at each position in the ZFPL and ZFPR binding sites are displayed as a logo for the group of 18-active off-target sites. Guanines at seven positions (red boxes) in the binding sites were absolutely conserved within these sequences, while they are more variable in the inactive sites (Supplementary Figure S5). (C) The distribution of the number of active (red) and inactive (blue) off-target sites as a function of the number of guanines preserved in each recognition sequence. (D) Each base in the binding site of the ZFPL and ZFPR was independently mutated to cytosine, which is not found at any position in either of the ZFP recognition sequences, and its influence on ZFP binding was assayed using B1H-based activity assay (38) at 1 mM 3-AT. This assay will detect only the most important positions for recognition, where a reduction in cell survival (plotted as the –log of surviving colonies) indicates a position important for recognition. All of the absolutely conserved guanines—indicated by an asterisk—are critical for activity.