Figure 2.
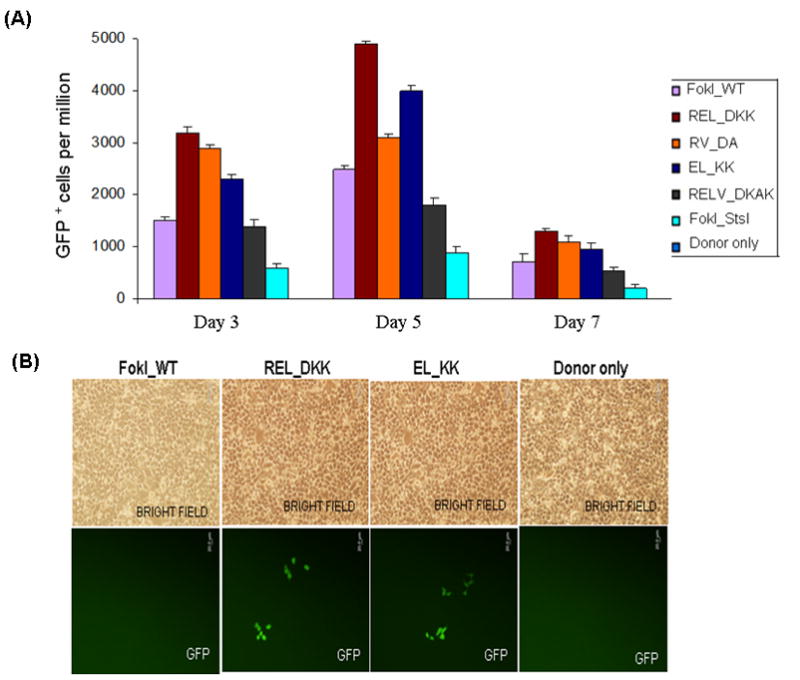
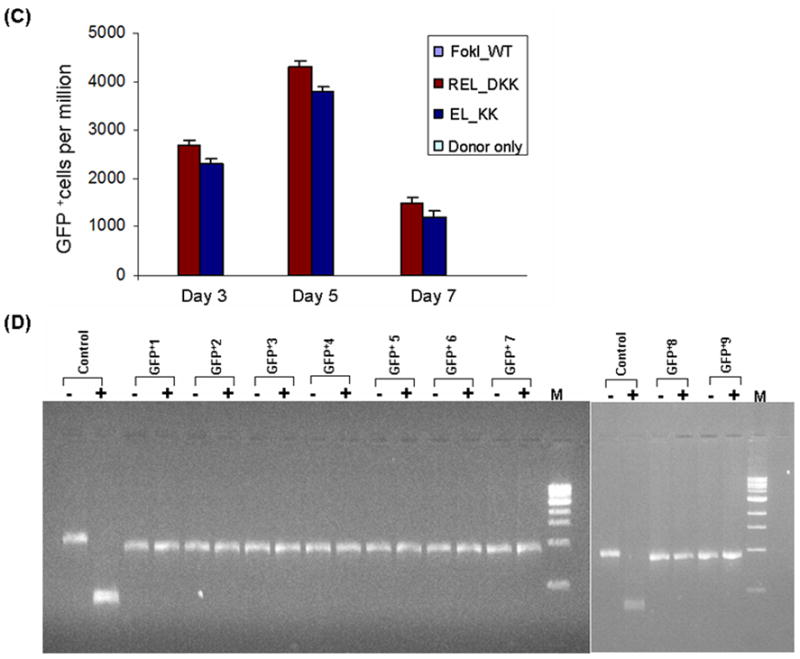
Testing the efficiency and efficacy of CCR5 3- and 4-finger ZFNs (generated by fusing the corresponding ZFPs to various obligate heterodimer variants of FokI nuclease domain) using the GFP gene targeting reporter system. HEK293 cells carrying a mutated eGFP reporter gene were transiently transfected with a donor plasmid carrying a fragment of wild-type GFP and plasmids expressing various 3- or 4-finger CCR5-specific ZFN constructs, using Lipofectamine 2000 as described elsewhere (21,27). Transfections of various obligate heterodimer variants and FokI_WT were performed one after another on the same day. After each transfection, the treated cells were split into 2 flasks. GFP positive cells in about 10,000 treated cells in each flask were determined by FACS and then normalized to one million treated cells. The difference between the two independent FACS readings is shown as error bars. A) Frequency of gene correction in HEK293 Flp-In cells of a chromosomal mutant GFP reporter disabled by insertion of the CCR5 ZFN target sequences using 4-finger ZFNs (28, 29). Quantitative FACS analyses of the GFP positive cells at 3, 5 and 7 days post-transfection with designer CCR5-specific 4-finger ZFNs (constructs carrying the FokI_WT and obligate heterodimer FokI nuclease domain variants) and donor plasmids. WT: wild type; EL_KK, 4-finger CCR5-ZFNs containing the FokI nuclease domain mutants reported by Miller et al., 2007 (32). RV_DA, 4-finger CCR5-ZFN constructs carrying the FokI nuclease domain mutants reported by Szczepek et al., 2007 (33). REL_DKK, RELV_DKAK and FokI_StsI, 4-finger CCR5-ZFN constructs carrying the FokI nuclease domain mutants that were generated at PBPL/JHU. B) Top panel: HEK293 Flp-In cells 5 days post-transfection as seen in brightfield; Bottom panel: GFP positive cells as seen 5 days post-transfection of HEK293 Flp-In cells with 3-finger CCR5-ZFN constructs (carrying Fok_WT, EL_KK or REL_DKK respectively) and donor plasmid. No GFP positive cells were seen with either donor alone or ZFNs containing FokI_WT nuclease domains and donor plasmid. C) FACS analyses of the frequency of gene correction in HEK293 Flp-In cells using 3-finger CCR5 ZFN constructs carrying FokI_WT, EL_KK, REL_DKK or donor alone, respectively. Results from two independent transfections, performed on different days, are shown in Figure S1; both transfections showed a similar trend of gene correction efficiencies for EL_KK and REL_DKK, respectively. The dose response curve using titrations of 3-finger ZFN expression plasmids, EL_KK and REL_DKK respectively, at constant donor plasmid, are shown in Figure S2. D) Analysis of the genotype of nine different individual GFP positive clones. Five days post-transfection with CCR5 3-finger ZFNs and the donor plasmids, GFP positive cells were sorted, serially diluted to get individual clones and grown. The genomic DNA was isolated from the GFP positive clones and the eGFP gene at the Flp-In locus was PCR amplified and digested with BstXI. The mutant eGFP gene has two BstXI sites, where the ZFN binding sites are inserted. Correction of the eGFP gene by homology-directed repair results in the loss of the BstXI sites. The PCR product size of the corrected eGFP gene is 930 bp as compared to 990 bp for the mutant gene. BstXI digestion of the mutant eGFP PCR product generates two bands: 450 bp and 540 bp, respectively. Lanes: Control, PCR product of the mutant eGFP gene from untransfected cells before (-) and after (+) digestion with BstXI; GFP+1–9, PCR products of 9 different individual clones obtained from GFP positive sorted cells before (-) and after (+) digestion with BstXI; M, 1 Kb ladder. All GFP positive cells are resistant to BstXI digestion, confirming ZFN-mediated eGFP gene correction in these cells.
