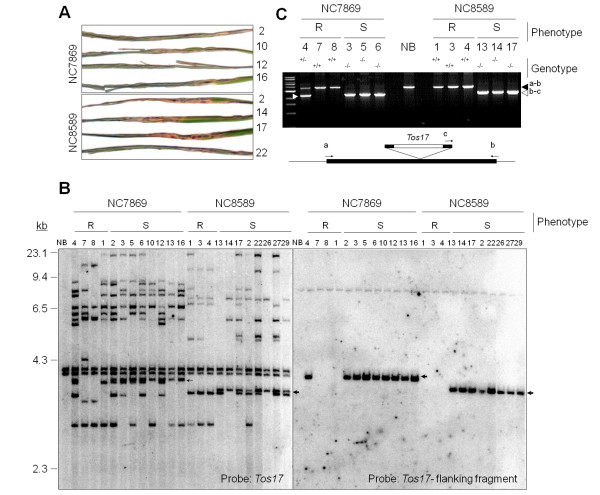
Figure 2.
Co-segregation analysis of mutant lines NC7869 and NC8589. (A) Representative phenotypes of the susceptible mutants that segregated in these lines. Numbers on the right of the photographs represent individual seedlings. (B) DNA gel blot analysis of XbaI-digested genomic DNA was carried out to investigate co-segregation between disease phenotypes and Tos17 insertions in individual plants of the NC7869 and NC8589 lines. Disease phenotypes are indicated at the top as R (resistant) and S (susceptible) to the blast fungus containing avrPish. Numbers at the top of each lane represent individual seedlings. NB served as a control for wild-type (resistant) seedlings. The blot was probed first with a 32P labeled Tos17 fragment (left) and then with a labeled fragment flanking the co-segregating Tos17 copy (right). The positions of size markers (λ DNA digested with HindIII) are shown on the left. Co-segregating bands are indicated by arrowheads. (C) Genotyping PCR analysis of genomic DNA from NC7869 and NC8589 plants exhibiting resistance (R) or susceptibility (S). The black triangle indicates the approximately 3.6 kb fragment that was amplified from the wild-type genome using primers at positions indicated by a and b on the diagram below the PCR gel. The white triangles indicate the fragments amplified using primers at positions b and c, designed to amplify the flanking sequence of Tos17. The genotype of each seedling is given at the top of each lane.