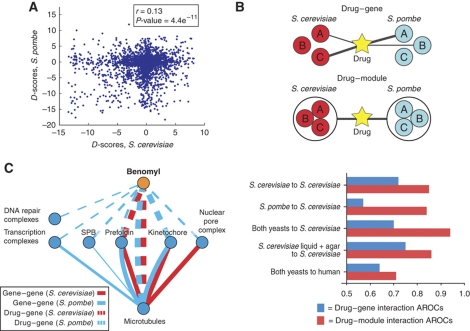
Figure 4.
Compound–module interactions are more conserved than compound–gene interactions. (A) A plot of the D-scores for the same compound–gene pairs in S. cerevisiae versus S. pombe. The level of observed conservation (r=0.13, P-value=4.4 × 10−11) is similar to the level of observed between gene–gene interaction partners (Roguev et al, 2008). (B) Use of the I-score to compare compound–gene versus compound–module interactions. We observe a higher conservation on the compound–module level by comparing the accuracy of using compound–gene interactions in S. pombe to predict compound–gene interactions in S. cerevisiae (AROC—area under the ROC curve=0.57) with using compound–module interactions for the same calculation (AROC=0.84). We observe that combining S. pombe compound–module interactions with S. cerevisiae compound–module interactions improves predictions over those made by either data set alone. Then we combine the S. cerevisiae data from Hillenmeyer et al (2008) with our S. cerevisiae data to carry out the same prediction, and we observe no significant improvement (compound–gene AROC=0.75, compound–module AROC=0.86). Finally, we use our cross-species data to predict small-molecule targets in human, and observe a predictive ability significantly higher than random (compound–gene AROC=0.64; compound–module AROC=0.72). (C) Comparison of compound–gene and gene–gene interactions. Compound–gene interactions are represented by dashed lines, gene–gene interactions are shown by solid lines. S. cerevisiae data are shown in red, and S. pombe data are shown in blue. Only highly significant interactions with P-value <0.005 are represented, and the line thickness is proportional to the significance of each interaction (−log(P-value)).