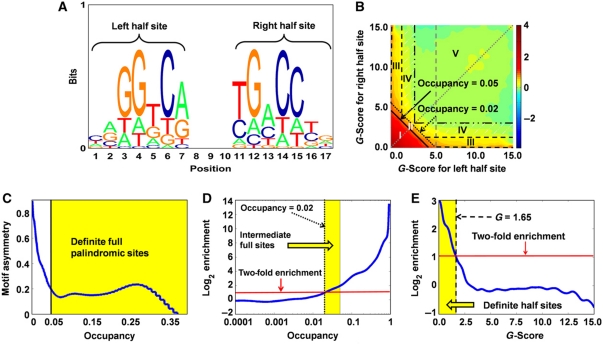
Figure 3.
De novo ER motif discovery and definition of five subcategories of ERα-binding site. (A) Sequence logo of palindromic ER motif deduced by Thermodynamic Modeling of chip-Seq (TherMoS) from MCF-7 ChIP-seq data. (B) Binding free energy of 17-mers relative to that of the consensus-binding sequence is decomposed into contributions (G-scores) from the left (GL; x axis) and right (GR; y axis) half sites. The plot shows log10-scale enrichment of G-score pairs in 100 bp regions centered on ChIP-seq peaks, relative to randomly chosen non-coding regions in the genome. Predicted probability of binding, i.e., occupancy  τ=2.3396. Area I: definite full site (occupancy>0.05); area II: intermediate full site (0.02<occupancy⩽0.05); area III: definite half site; area IV: intermediate half site; area V: no ERE. (C) Occupancy threshold for definite full (palindromic) sites was defined as the value below which GL and GR become anti-correlated, i.e., asymmetric. (D) Occupancy threshold for intermediate sites corresponds to the point on the dotted diagonal line in B wherein the enrichment is twofold. The same point on the diagonal line is used to define the G-score threshold for intermediate half sites (dashdot line in B). (E) G-score threshold for definite half sites is the point on the dashed vertical line in B where the enrichment is twofold.
τ=2.3396. Area I: definite full site (occupancy>0.05); area II: intermediate full site (0.02<occupancy⩽0.05); area III: definite half site; area IV: intermediate half site; area V: no ERE. (C) Occupancy threshold for definite full (palindromic) sites was defined as the value below which GL and GR become anti-correlated, i.e., asymmetric. (D) Occupancy threshold for intermediate sites corresponds to the point on the dotted diagonal line in B wherein the enrichment is twofold. The same point on the diagonal line is used to define the G-score threshold for intermediate half sites (dashdot line in B). (E) G-score threshold for definite half sites is the point on the dashed vertical line in B where the enrichment is twofold.