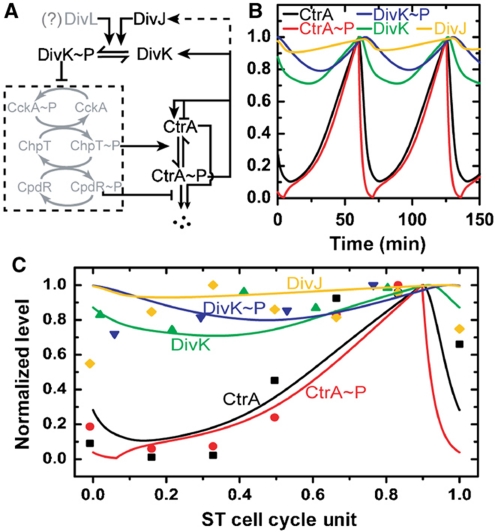
Figure 3.
Deterministic simulation of C. crescentus ST cell cycle oscillatory period. (A) Simplified regulatory network for ST cell cycle. Regulation of CtrA oscillation is modeled by a five-component protein interaction network (see Materials and methods section for details). The model only includes the components shown in black (i.e. the gray-shaded part of the network is simplified). The dash box indicates a phosphorelay system that is approximated as a binary switch controlled by the DivK∼P level. (B) Protein concentrations (normalized) trajectories over about two cell cycle periods. The model is described by coupled differential equations and is solved to obtain trajectories as shown. (C) Comparison of simulation with experimental data. Experimental protein levels (solid symbols) are extracted from published immunoblot data for the five components included in the model (CtrA (Holtzendorff et al, 2004), CtrA∼P (Jacobs et al, 2003), DivK (Jacobs et al, 2001), DivK∼P (Jacobs et al, 2001), and DivJ (Wheeler and Shapiro, 1999)). The cell cycle periods are normalized to 1. ST cell data are then taken from 15 to 100% of the SW cell data (∼15% of time for SW-ST transition (Keiler and Shapiro, 2003)) and the time is rescaled by 0.85 to yield ST cell cycle unit (0–1).