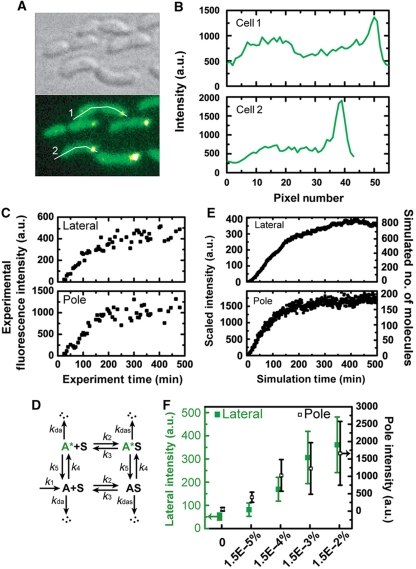
Figure 7.
Single-cell fluorescence localization, kinetics of DivJ–EGFP and model simulation. (A) C. crescentus cells in microfluidic culture expressing DivJ–EGFP. Bright-field DIC image (upper left) and a false-color fluorescence image from the same area (lower left) for divJ∷Tn5 (ΔdivJ) cells expressing divJ-egfp from the xylX chromosomal locus for 0.015% xylose induction. (B) Spatial profile of EGFP fluorescence. The plots are the fluorescence intensity profiles along the white lines indicated in (A) for cell #1 (upper) and cell #2 (lower). The horizontal coordinate represents the distance from the leftmost of each line in pixels. (C) Sample single-cell temporal fluorescence intensity traces obtained for 0.015% xylose for the same mutant construct. (D) Kinetic model where the species corresponding to experimental observables are A* (lateral membrane-bound) and A*S (pole localized). (E) Time traces for A* and A*S from simulation. The numbers of molecules are simulated with k1=0.37 (Supplementary Table VII). The simulated molecule numbers are scaled as intensities for comparison with experimental trajectories (see Supplementary information for details). (F) Steady-state fluorescence intensities obtained for different inducer concentrations. Cells in the microfluidic channel were induced with different levels of xylose for 6 h and their intensities were characterized, except for the zero xylose condition in which the intensities were characterized before induction. Lateral membrane intensities (green solid squares) and stalked pole intensities (black open squares) are associated with the left and right y-axis, respectively. Error bars represent s.d. (n=23–29).