Abstract
Identifying factors influencing transposable element activity is essential for understanding how these elements impact genomes and their evolution as well as for fully exploiting them as functional genomics tools and gene-therapy vectors. Using a genetics-based approach, the influence of genomic position on piggyBac mobility in Drosophila melanogaster was assessed while controlling for element structure, genetic background, and transposase concentration. The mobility of piggyBac elements varied over more than two orders of magnitude solely as a result of their locations within the genome. The influence of genomic position on element activities was independent of factors resulting in position-dependent transgene expression (“position effects”). Elements could be relocated to new genomic locations without altering their activity if ≥500 bp of genomic DNA originally flanking the element was also relocated. Local intrinsic factors within the neighboring DNA that determined the activity of piggyBac elements were portable not only within the genome but also when elements were moved to plasmids. The predicted bendability of the first 50 bp flanking the 5′ and 3′ termini of piggyBac elements could account for 60% of the variance in position-dependent activity observed among elements. These results are significant because positional influences on transposable element activities will impact patterns of accumulation of elements within genomes. Manipulating and controlling the local sequence context of piggyBac elements could be a powerful, novel way of optimizing gene vector activity.
TRANSPOSABLE elements are ubiquitous components of genomes that have played and continue to play a significant role in shaping the size and organization of genomes and contribute to genetic variation within genomes upon which natural selection can act (Kidwell and Lisch 2001; Kidwell 2002; Pritham 2009; Rebollo et al. 2010). The contribution of transposable elements to the dynamic processes that shape genomes and the biological significance of these elements to the organisms that harbor them are due largely to the excision and/or transposition activities of the elements. Understanding the factors that influence transposable element activity, therefore, is important for understanding how transposable elements exert their influences on the genomes in which they are found.
Understanding transposable element movement and regulation is of considerable practical importance because they have become valuable functional-genomics tools that aid in the identification, isolation, and analysis of genes. Transposable elements are also popular genetic platforms from which gene vectors are constructed for the purposes of creating transgenic cells and organisms (Ivics et al. 2009; VandenDriessche et al. 2009; Bouuaert and Chalmers 2010; Carlson and Fahrenkrug 2010; Munoz-Lopez and Garcia-Perez 2010). Recently, the practical significance of transposable elements has been extended to include clinical applications where these elements are being developed and used as human gene therapy vectors to deliver genes to cells and tissues for the purpose of improving the health of patients (Ivics and Izsvak 2006; VandenDriessche et al. 2009; Bouuaert and Chalmers 2010).
Although transposable elements are widely distributed and abundant, in most genomes few elements are usually active because most have become defective and dysfunctional as a result of mutations acquired over time and during the excision/transposition/repair process. Not only are active elements rare, relative to the total number of elements within most genomes, but also the rates of movement of those that are active are highly variable. For example, in Drosophila melanogaster the number of insertions per element per generation has been estimated from 2 × 10−1 to 3 × 10−4 (Robertson et al. 1988; Engels 1989). There are many factors known to influence element activity, including the concentration of element-specific transposase, the size, structure, content, and organization of the element, and the presence of regulatory proteins and/or small RNAs (Feschotte and Pritham 2007; Malone and Hannon 2009; Obbard et al. 2009). Chromatin structure and organization are expected to influence the movement and distribution of transposable elements within genomes although this aspect of transposable element movement remains largely unexplored. The distinctly nonrandom distributions of transposable elements within genomes probably reflects, in part, constraints on the ability of transposases to interact with DNA substrates when they are part of chromatin structures, although direct evidence for the importance of chromatin in modulating transposable element movement remains largely absent (Pryciak and Varmus 1992; Muller and Varmus 1994; Pruss et al. 1994; Slotkin and Martienssen 2007). The clustering of transposable element insertions around the 5′ region of genes, as is the case with P elements in D. melanogaster and Mu in Zea mays, has been taken as evidence for the preference of these two elements for integrating into DNA associated with open chromatin configurations (Liao et al. 2000; Liu et al. 2009). However, the distribution of class II elements within genomes is also a function of the remobilization activity of elements, i.e., the tendency of elements to excise and/or transpose from particular sites within the genome. While little is known about how element activity is influenced by genomic position, there is some evidence indicating that the remobilization potential of elements is non-uniformly distributed across the genome.
Engels (1989) reported that a nonautonomous P element inserted at cytological position 17B in the genome of D. melanogaster displayed very low frequencies of transposition and excision even when high levels of functional P-element transposase were present. When a transposition event involving this element eventually was recovered, resulting in the insertion of the element in a new genomic position (22A), the newly relocated element transposed subsequently at frequencies considered typical of P elements (Engels 1989). Other P elements at different positions within the genome and resulting from the transposition of the element at 22A were also readily remobilized. Engels (1989) suggested that the stability of the original P element at cytological position 17B was due in some way to its position within the genome. Robertson et al. (1988) reported excision and transposition frequencies of a number of P elements in the presence of a constant source of transposase and found the rates of movement to be highly variable. Although there was variation in the structures of the elements tested in some cases, the variability in element movement was largely a function of the elements' positions within the genome. Berg and Spradling (1991), while investigating some of the factors that affect the mobility of P elements in D. melanogaster, found that not only the structure of elements but also the chromosomal position of the elements was an important factor affecting transposition frequency (Berg and Spradling 1991). They found that two identical P elements at cytological positions 7D and 9AB had a sixfold difference in transposition activity in the presence of identical levels of transposase. Similarly, Ds elements in different locations within the genome of Arabidopsis thaliana were also found to have different rates of excision/transposition in the presence of fixed sources of transposase (Bancroft and Dean 1993; Smith et al. 1996). Bancroft and Dean (1993) reported that a 10-fold difference in the excision rates of two identical Ds elements could be attributed solely to where the elements were located within the genome (Bancroft and Dean 1993). Lisch et al. reported that both MuDR-1 excision and duplicative transposition rates were influenced by chromosomal position in maize (1995). They measured a threefold difference in the frequency of duplicative transpositions between two unlinked elements and report as unpublished observations that elements at other positions had an even greater effect on transposition rates. Kitamura et al. (2001) made similar observations of the Tam3 element in Antirrhinum majus (Kitamura et al. 2001). The frequency of movement of nearly identical Tam3 elements varied as a function of genomic position, but in this case element activity seemed correlated to the degree of methylation among the elements. In none of these systems was it known what the proximal cause for the observed differences in element mobilities was, but differences in the conformation of local chromatin were thought to influence the accessibility of critical cis-essential factors required for excision/transposition (Engels 1989; Bancroft and Dean 1993).
Here we examine the role of chromosomal position on piggyBac activity in D. melanogaster and show that, in an isogenic genetic background and a constant source of transposase, piggyBac activity varies over two orders of magnitude. Position effects influencing piggyBac activity are distinct from and independent of position effects affecting transgene expression from within the element. We show that the observed variation in element mobility resulted from properties intrinsic to the local chromosomal DNA extending no further than 500 bp from the ends of the piggyBac element. These intrinsic properties are portable to new chromosomal locations as well as to plasmids. The level of piggyBac exision/transposition activities within the genome of D. melanogaster is strongly associated with the predicted bendability of the DNA immediately adjacent to the 5′ and 3′ terminal inverted repeats. These results are significant because they reveal the importance of local genomic features in transposable element movement and provide a rationale by which piggyBac-based gene vectors could be modified or designed for optimized transposition activity.
MATERIALS AND METHODS
Fly stocks:
D. melanogaster lines were obtained from the Bloomington Drosophila Stock Center at Indiana University and the Exelixis Collection at the Harvard Medical School and maintained at 25° on a standard cornmeal/yeast-based diet. The following fly stocks were used (stock number or line ID followed by genotype):
1-Canton-S: wild type
3605-w1118: a homozygous recessive spontaneous loss-of-function mutation in the white gene resulting in white eyes containing < 1% of normal pigmentation levels.
8285-w1118 CyO, P{Tub - PBac\T}2/wgSp-1: multiple copies of a P element containing the piggyBac transposase open reading frame under the regulatory control of the strong constitutive promoter from the D. melanogaster tubulin gene on the CyO balancer chromosome.
c00169-PBac{PB}CG4061c00169; c00178-PBac{PB}c00178; c00404-PBac{PB}CG15335c00404; e00747-PBac{RB}e00747; c01352-PBac{PB}c01352; c01384-PBac{PB}CG32549c01384; c01423-PBac{PB}c01423; c01465-PBac{PB}c01465; c02335-PBac{PB}CG1677c02335; e02382-w1118 PBac{RB}CG2247e02382; c03178-PBac{PB}CG3600c03178; f03565-PBac{WH}pod1f03565; c03764-PBac{PB}c03764; c04386-PBac{PB}Ptp10Dc04386; c04426-PBac{PB}AlstRc04426; c04432-PBac{PB}c04432; c05494-PBac{PB}c05494; c05577-PBac{PB}CG3600c05577; c05745-PBac{PB}CG3638c05745; c06451-PBac{PB}C3Gc06451; f07297-PBac{WH}mei-217f07297: piggyBac-containing lines with a single nonautonomous element containing the mini-white gene (w+mC) (Thibault et al. 2004). All insertions are on the X chromosome with the exception of c00747, which is on 3L.
24483-y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-51D: contains a ϕC31 attP site on the right arm of chromosome 2 and the ϕC31 integrase gene under the regulatory control of the promoter from the vasa gene on the X chromosome (Bischof et al. 2007).
23648-P{ry+t7.2=hsp70-flp}1, y1 w*; M{3xP3-RFP.attP}ZH-86Fb; M{vas-int.B}ZH-102D: contains a ϕC31 attP site on the right arm of chromosome 3 and the ϕC31integrase gene under the regulatory control of the promoter from the vasa gene on chromosome 4 (Bischof et al. 2007).
24481-y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP'}ZH-22A: contains a ϕC31 attP site on the left arm of chromosome 2 and the ϕC31 integrase gene under the regulatory control of the promoter from the vasa gene on the X chromosome (Bischof et al. 2007).
24485-y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP'}ZH-68E: contains a ϕC31 attP site on the right arm of chromosome 2 and the ϕC31 integrase gene under the regulatory control of the promoter from the vasa gene on the X chromosome (Bischof et al. 2007). See Table S1 for additional details.
Molecular biology:
The piggyBac elements from selected lines and with various amounts of flanking genomic DNA were amplified using the polymerase chain reaction (PCR) with element-specific primers (see Table S2 for the details of the primers used). Amplifications were performed under the following conditions: 1× Phusion buffer (Finnzymes, Woburn, MA), 0.2 mm of each dNTP, 0.1 μm primer forward, 0.1 μm primer reverse, 1 unit Phusion DNA polymerase (Finnzymes) and 5–20 ng of genomic DNA. The PCR reactions were performed under the following conditions: 3 min at 98°; 5 cycles of 15 sec at 98°, 30 sec at 62°, and 5 min at 72°; 35 cycles of 15 sec at 98°, 30 sec at 68°, and 5 min at 72°; 10 min at 72°. The PCR fragments were ligated into pCR-Blunt II TOPO vectors according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). This resulted in plasmids that contained piggyBac elements marked with mini-white and flanked by various amounts of genomic DNA and containing a unique AscI restriction site in one of the PCR primers.
The ϕC31 attB site was amplified from the plasmid pUASTattB (Bischof et al. 2007) using the PCR under the following conditions: 1× ThermoPol buffer (New England Biolabs, Ipswich, MA), 0.4 mm of each dNTP, 0.2 μm primer AttBFor, 0.2 μm primer AttBRev, 5 units Taq DNA polymerase (New England Biolabs), and 5–20 ng of template DNA. The PCR reactions were carried out under the following conditions: 5 min at 95°; 25 cycles of 30 sec at 95°, 30 sec at 56°, and 30 sec at 72°; 5 min at 72°. The resulting fragments were ligated into a pCR2.1-TOPO vector according to the manufacturer's recommendations (Invitrogen). Because the forward and reverse primers contained AscI restriction sites, the ∼300-bp attB-containing fragment could be removed from the pCR2.1plasmid by AscI digestion (Table S2).
ϕC31 attB sites were added to pCR-Blunt II plasmids containing isolated piggyBac elements by digesting the plasmids with AscI, dephosphorylating the terminal nucleotides of the linearized plasmids, and ligating the resulting DNA to 300-bp attB-containing fragments obtained by digesting the pCR2.1-TOPO with the ϕC31 attB site with AscI.
Germline transformation of D. melanogaster:
Subcloned piggyBac elements on ϕC31attB-containing plasmids were introduced into ϕC31attP-containing lines of D. melanogaster that were expressing ϕC31 integrase in the germline as described (Bischof et al. 2007). Preblastoderm embryos were injected with plasmids at a concentration of 0.25 mg/ml, and individual adults arising from these embryos were out-crossed to flies of the opposite sex from line 3605 (w1118). Red-eyed transgenic progeny of these crosses were used to establish homozygous lines.
The subcloned piggyBac elements from lines 02382 and 05577 were also used as gene vectors to create transgenic D. melanogaster via piggyBac transposition. A mixture of pCR-Blunt II-02382+1kb, pCR-Blunt II-05577+1kb, pBac[3xP3-DsRed] (Horn and Wimmer 2000), and phsp-pBac (Handler and Harrell 1999), each at 0.2 mg/ml, was injected into preblastderm embryos of the line 3605 (w1118), and the resulting adults were individually crossed to uninjected flies of the opposite sex from line 3605 (w1118). Transgenic progeny from these crosses were recognized by the presence of red-pigmented eyes.
Measuring piggyBac activity and somatic activity:
Males containing the mini-white gene-containing piggyBac element to be mobilized were crossed en mass with virgin females of transposase-expressing line 8285 (w1118 CyO, P{Tub-PBac\T}2/wgSp-1). Curly winged, red-eyed progeny were examined for the presence of clones of ommatidia with no pigmentation (arising from piggyBac excision) or with levels of pigmentation more or less than the parental line [arising from piggyBac transposition and different levels of mini-white expression as a result of position effects (Levis et al. 1985)]. Activity was expressed as the proportion of curly-winged, red-eyed progeny with eye-color mosaicism. Quantitative analysis of the data was performed using the R statistical software (R Development Core Team 2010). P-values resulting from pairwise ANOVA have been adjusted according to the Tukey HSD method.
Measuring piggyBac activity and germline activity:
Females containing an X-linked, mini-white gene-containing piggyBac element were crossed en mass with males of transposase-expressing line 8285 (w1118 CyO, P{Tub-PBac\T}2/wgSp-1). Fifty curly winged, red-eyed male progeny were crossed individually to 5–10 virgin females from line 3605 (w1118), and the presence of red-eyed male progeny arising from this cross was noted. Because the piggyBac element was X-linked, red-eyed male progeny were not expected unless the element was transpositionally active, in which case transposition to an autosomal chromosome would result in exceptional male progeny containing the transposable element and transgene. Germline activity was expressed as the proportion of germlines of curly winged, red-eyed males that produced exceptional red-eyed male progeny.
Quantification of mini-white transgene expression:
Ten heads were collected from 4-to 5-day-old adult females homozygous for the mini-white gene-containing piggyBac elements and wild-type flies from the Canton-S line. Fresh tissue was preserved immediately in 50 μl of RNAlater RNA Stabilization Reagent (Ambion, Austin, TX). Three collections were performed for each line of flies analyzed. Total RNA was isolated from homogenized tissue using the RNeasy Plus Mini Kit (QIAGEN, Valencia, CA) and used as template for a two-step reverse transcription–quantitative PCR (RT–qPCR) reaction utilizing Access RT–PCR System reagents (Promega, Madison, WI).
First-strand cDNA synthesis:
Separate 35-μl first-strand cDNA reaction mixtures were prepared for analyzing transcript abundance of the control gene, Ribosomal protein 49 (RP49), and the target gene, white, using 25 ng of total RNA as template, nuclease-free water, and 1.0 μl of 2 μm reverse primers (Table S2). The template-primer mixtures were heated to 65° for 10 min in a thermal cycler and cooled on ice for 5 min. A 16-μl aliquot of Promega's Access RT–PCR reagents in a master-mix cocktail—consisting of 1× avian myeloblastosis virus (AMV) buffer, 0.2 mm of each dNTP, 1 mm MgSO4, 0.1 unit AMV reverse transcriptase, and 1.6 units RNase Inhibitor (New England Biolabs)—was added to each template-primer mixture to make a 50-μl reaction volume. The first-strand cDNA synthesis reaction was performed at 45° for 45 min, and reverse transcriptase was inactivated by heating the reaction to 94° for 2 min.
Real-time SybrGreen qPCR:
The qPCR reaction mixture using Access RT–PCR system reagents (Promega) was assembled so that the final concentration of the components consisted of 1× reaction buffer, 0.2 mm of each dNTP, 1 mm MgSO4, 0.2 μm forward primer (Table S2), 0.2 μm reverse primer (Table S2), 2.5 units Tfl DNA polymerase, and 1 μl of a 1/30,000 dilution of SybrGreen (Fluka, Buchs, Switzerland) in 100% DMSO. The reaction mixture was assembled as a master mix to which was added 1 μg of the appropriate cDNA, which was then subdivided to accommodate the analysis of three technical replicates of the white and RP49 genes. Quantitative real time PCR was performed with the 7300 Real-Time PCR System (Applied Biosystems, Carlsbad, CA) using the following thermal cycling conditions: 5 min at 94° followed by 40 cycles of 30 sec at 94°, 30 sec at 61°, and 20 sec at 72°. Following the PCR, the reactions were analyzed for the presence of primer dimers by treating them for 15 sec at 95°, ramping to 30 sec at 60°, and ramping to 95° for 15 sec. The specificity of the amplification was also checked by analyzing the PCR products on 1.5% agarose gels. The resulting data were analyzed using the comparative CT method (Schmittgen and Livak 2008). Thresholds and baselines were set manually; thresholds were chosen within the exponential region of the semilog PCR amplification plots.
DNA bendability:
For a flanking sequence of interest, the bendability/curvature propensity plot was calculated beginning at base −15 from the end of the element to 200 bp from the terminal inverted repeats of the piggyBac element with the bend.it server (http://hydra.icgeb.trieste.it/dna/bend_it.html), using the DNaseI-based bendability parameters of Brukner et al. (1995) and the consensus bendability scale of Gabrielian and Pongor (1996). The mean bendability for the sequence of interest was calculated, and the association between mean sequence bendability and element activity was analyzed using linear regression models calculated using the method of ordinary least squares. Mean sequence bendability was calculated over varying lengths of flanking DNA. As controls, 25 sets of random sequences were generated on the basis of the combined nucleotide abundance found in the 19, 5′ and 3′ 200-bp flanking sequences used in this analysis. The randomized sequences were arbitrarily associated with known element activities, and the coefficient of determination (r2) was calculated using linear models as before. The data from the analysis of 25 data sets consisting of randomized flanking sequences were summarized by calculating the mean and standard deviation of the coefficients of determination. All calculations associated with this analysis were performed on programs written and executed in Mathematica 7 (Wolfram Media, Champaign, IL).
piggyBac integration-site analysis:
Using FlyBase version FB2010_07 (Tweedie et al. 2009) piggyBac integration events were identified in the genome of D. melanogaster and those sites in which integration events occurred two or more times in the same orientation were considered “hot spots” and retained for further analysis. The chromosomal DNA sequence flanking the 5′ terminal-inverted-repeat of piggyBac elements inserted at integration “hot spots” were used for calculating DNA bendability as described above.
RESULTS
Genomic location greatly influences transposable element activity:
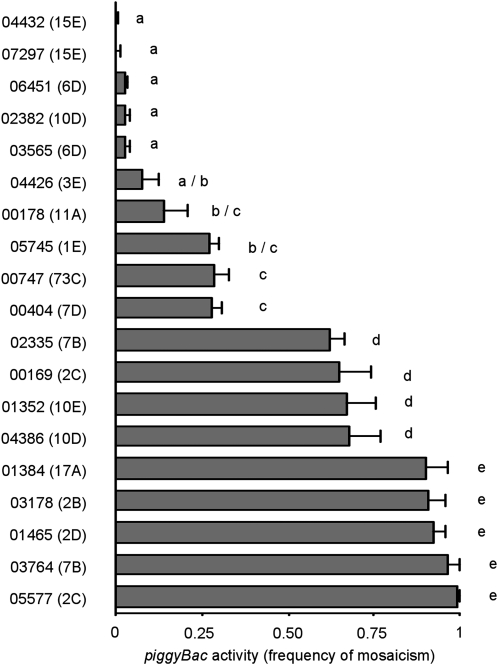
We sampled a collection of transgenic D. melanogaster in which each individual contained a single piggyBac element genetically marked with the mini-white gene in an isogenic genetic background (Thibault et al. 2004) and measured somatic (and in two cases germline) activities of each element. The remobilization activity of the piggyBac elements tested in this study was completely dependent upon the presence of piggyBac transposase but varied greatly, with some elements being almost immobile under these experimental conditions while others were extremely active. Because the elements tested were identical or nearly identical (Thibault et al. 2004) in an isogenic background with an invariable source of transposase, the differences in activities could be attributed entirely to the effects of the elements' positions within the genome. The activity of piggyBac appears to be continuously variable, and we estimate that the differences in activity levels between the least active and most active elements in this study were at least 100-fold (Figure 1, Table S3). In addition to measuring the activity of piggyBac in somatic cells, the germline transposition activity of the elements in lines 02382 and 05577 was measured. In both 02382 and 05577, the amount of germline activity reflected the observed level of somatic activity with the relatively inactive element in line 02382 producing germline transposition events in only 4% of the germlines tested (ngermlines tested = 50) while the somatically active element in line 05577 resulted in transposition in 80% of the germlines tested (ngermlines tested = 50).
Figure 1.—
Activity of piggyBac elements in different lines. The mean proportion (±SD; n = 3) of flies that are heterozygous for a piggyBac element and a piggyBac transposase gene with mosaic eye color (element activity) is shown for each of the lines used in this study. A one-way analysis of variance (ANOVA) was performed to compare the effect of element location on element activity. There was a significant effect of element location on element activity: F[18,38]=149.66, P < 0.0001. Post-hoc comparisons of all pairs of means using Tukey's HSD test indicated which elements had significantly different activities (P < 0.05). Elements with significantly different means are indicated with different lower case letters.
Position effects on transposable element activity and transgene expression are distinct:
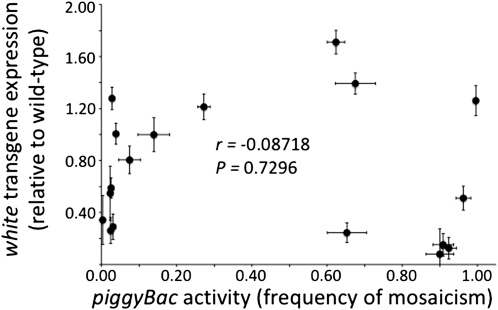
The level of expression of the mini-white transgene within the piggyBac element was measured using qRT–PCR on similarly aged females homozygous for the piggyBac element under conditions in which the element is not active. The gene expression levels from the mini-white gene in the piggyBac elements were not correlated with the transposition activities of the elements (Pearson's correlation coefficient, r[16]=−0.08718, F[1,16]=0.1237, P = 0.7296) (Figure 2). piggyBac elements with high rates of remobilization could have high or low levels of transgene expression as could piggyBac elements with low rates of transposition activity.
Figure 2.—
Correlation between transgene expression and piggyBac activity. Mean white-transgene expression (± standard error; n = 3) in females and mean element activity (± standard error; n = 3) were plotted, and Pearson's correlation coefficient was calculated. Transgene expression was measured by qRT–PCR and expressed as a proportion of white gene expression in similarly aged Canton-S (wild-type) females. Element activity is expressed as the proportion of flies from a test cross with mosaic eye color. No correlation was found (r = −0.08718, P = 0.7296).
Intrinsic local properties of element-flanking DNA determine element activity:
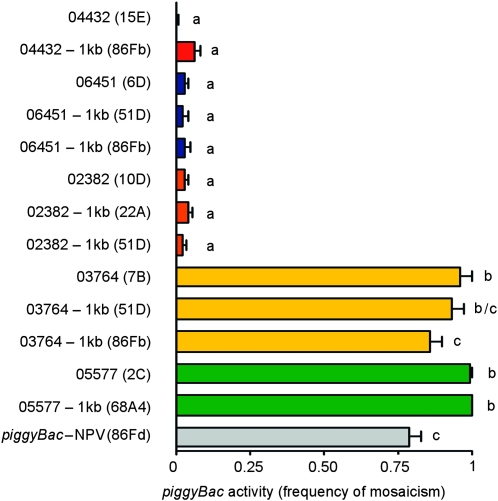
piggyBac elements with known rates of remobilization along with 1 kb of genomic DNA immediately flanking the 5′ and 3′ terminal inverted repeats were amplified using the PCR, cloned into a plasmid containing the attB site of the bacteriophage C31 (ϕC31), and reintegrated into the genome of D. melanogaster containing the attP site of ϕC31 at known locations (Groth et al. 2004; Bischof et al. 2007; Fish et al. 2007) (Figure 3; Table 1). The transposition/excision activities of the relocated transposable elements were measured (Figure 4). Transposable elements relocated to new chromosomal positions along with 1 kb of the original flanking chromosomal DNA had activity levels that were not significantly different from the transposable elements in their initial chromosomal locations, with one exception (Figure 4, Table S3). Elements that had very low rates of remobilization from their original chromosomal location (e.g., lines 06451, 04432, 02382) also had similarly low rates of remobilization when they were relocated to one of three positions within the genome (Figure 4, Table S3). Likewise, when elements with very high rates of remobilization (e.g., lines 03764, 05577) were transplanted to the same positions within the genome, they largely retained their original remobilization characteristics (Figure 4, Table S3). In only one case did the relocated element have a remobilization rate statistically different from the element in its original location. When the element in line 03764 was relocated to cytological position 86Fb, it had a somewhat, but statistically significant, lower level of activity 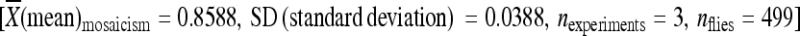 than the element at the original location (
than the element at the original location (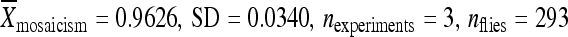 ) (Figure 4, Table S3).
) (Figure 4, Table S3).
Figure 3.—
Structure of local chromosomal environments of relocated piggyBac elements. Transgenic D. melanogaster flies were created by site-specific recombination between ϕC31attB-containing plasmids (dashed line) that have the mini-white gene-containing piggyBac element being relocated (blue arrow with white box) with varying amounts of genomic DNA from the original integration site (orange, yellow, red, green, and blue rectangles) and ϕC31attP sites (light blue and gray rectangle) that were placed in the genome of D. melanogaster (thin black line) using a Mos1 gene vector with a 3xP3DsRed marker gene (thick black line with white box) (Bischof et al. 2007). The element labeled “piggyBac” was the element from line 02382 without any neighboring chromosomal DNA. The element labeled “piggyBac-NPV” was the element from the plasmid PB{RB} with 1 kb of DNA flanking each terminal inverted repeat (gray rectangle) (Thibault et al. 2004). PB{RB} was used to create most of the lines used in this study. The first 150 bp of the DNA flanking the 5′ and 3′ terminal inverted repeats of the element in plasmid PB{RB} originated from the nuclear polyhedrosis virus from which piggyBac was originally isolated (Cary et al. 1989). The features represented in this diagram were not drawn to scale.
TABLE 1.
Transgenic lines obtained
| Name | Chromosome | Cytology | Construction | Left DNA (bp)a | Right DNA (bp)a |
|---|---|---|---|---|---|
| piggyBac (86Fb) | 3R | 86Fb | piggyBac | 0 | 0 |
| piggyBac (51D) | 2R | 51D | piggyBac | 0 | 0 |
| piggyBac–NPV(86Fb) | 3R | 86Fb | piggyBac-NPV | 966b | 1084b |
| 02382 – 1 kb (51D) | 2R | 51D | 02382 – 1 kb | 1009 | 997 |
| 02382 – 1 kb (22D) | 2L | 22D | 02382 – 1 kb | 1009 | 997 |
| 02382 – 500 bp (86Fb) | 3R | 86Fb | 02382 – 500 bp | 498 | 520 |
| 02382 – 250 bp (86Fb) | 3R | 86Fb | 02382 – 250 bp | 264 | 328 |
| 02382 – 20 bp (86Fb) | 3R | 86Fb | 02382 – 20 bp | 21 | 20 |
| 03764 – 1 kb (86Fb) | 3R | 86Fb | 03764 – 1 kb | 935 | 1083 |
| 03764 – 1 kb (51D) | 2R | 51D | 03764 – 1 kb | 935 | 1083 |
| 04432 – 1 kb (86Fb) | 3R | 86Fb | 04432 – 1 kb | 1027 | 973 |
| 04432 – 200 bp (86Fb) | 3R | 86Fb | 04432 – 200 bp | 210 | 207 |
| 05577 – 1 kb (68A4) | 3L | 68A4 | 05577 – 1 kb | 1032 | 1013 |
| 06451 – 1 kb (51D) | 2R | 51D | 06451 – 1 kb | 996 | 899 |
| 06451 – 1 kb (86Fb) | 3R | 86Fb | 06451 – 1 kb | 996 | 899 |
| 06451 – 200 bp (86Fb) | 3R | 86Fb | 06451 – 200 bp | 219 | 223 |
Amount of neighboring chromosomal DNA from original integration site accompanying piggyBac.
Amount of plasmid DNA from nuclear polyhedrosis virus sequence accompanying piggyBac.
Figure 4.—
Activity of relocated piggyBac elements with 1 kb of neighboring chromosomal DNA. Activity (mean ± SD; n = 3) is expressed as the proportion of flies with mosaic eyes in a remobilization testcross. The cytological positions of the transplanted elements are indicated in parentheses. A one-way ANOVA was performed to compare the effect of element location on element activity. There was a significant effect of element location on element activity: F[13,28]=1213.6, P < 0.0001. Post-hoc comparisons of all pairs of means using Tukey's HSD test indicated which elements had significantly different activities (P < 0.05). Elements with significantly different means are indicated with different lower case letters.
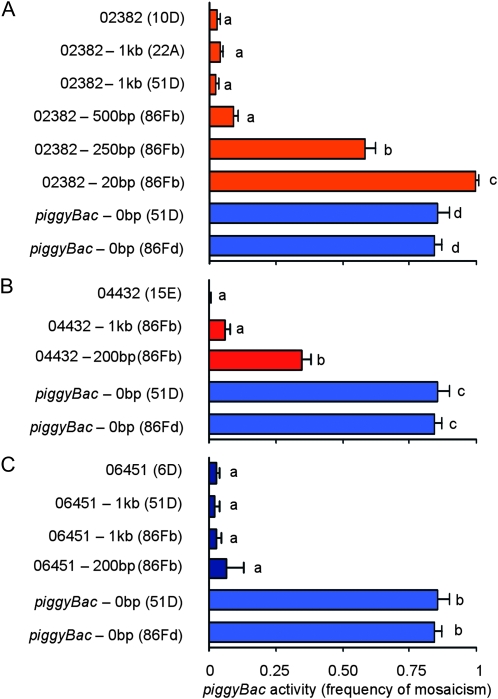
The results reported here demonstrate that local, intrinsic properties of the element-flanking DNA influence the ability of piggyBac transposable elements to be remobilized and that all of the information responsible for position-dependent variation in transposable element activity lies within 1 kb from the terminal inverted repeats of the element. To estimate the minimum length of flanking DNA that can be relocated with piggyBac elements without altering the remobilization behavior of the element, we repeated the relocation experiment using the element in line 02382 that had a low rate of remobilization in its original chromosomal position and included 500, 250, 20, and 0 bp of the original chromosomal DNA (Figure 5). When the element was relocated without any of the original flanking chromosomal DNA (i.e., 0 bp of the original chromosomal DNA), the element was then flanked by sequences from the PCR-cloning vector used for cloning the element (Figure 3, Table 1). Under these conditions, the element became highly active, yielding ∼ 85% mosaic individuals during remobilization assays (at 51D: 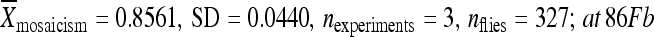 :
: 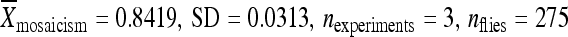 ) (Figure 5A). Relocating the element with 500 bp of the original chromosomal DNA to position 86Fb resulted in no significant change in the element's activity with the element remaining inactive (
) (Figure 5A). Relocating the element with 500 bp of the original chromosomal DNA to position 86Fb resulted in no significant change in the element's activity with the element remaining inactive (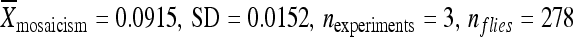 ) relative to the element in its original genomic position (
) relative to the element in its original genomic position (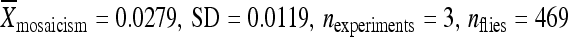 ) (Figure 5A). When only 250 bp of chromosomal DNA was included with the relocated element, the element was only moderately active (
) (Figure 5A). When only 250 bp of chromosomal DNA was included with the relocated element, the element was only moderately active (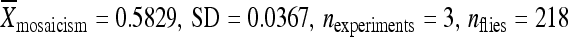 ) (Figure 5A). Relocating the element with only 20 bp of the original chromosomal integration site to cytological position 86Fb resulted in an element with high remobilization activity (
) (Figure 5A). Relocating the element with only 20 bp of the original chromosomal integration site to cytological position 86Fb resulted in an element with high remobilization activity (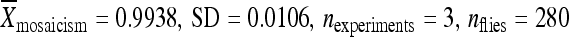 ) (Figure 5A). This activity was statistically different from the activity of the element that completely lacked any chromosomal DNA from the insertion site of the element from line 02382 (Figure 5A).
) (Figure 5A). This activity was statistically different from the activity of the element that completely lacked any chromosomal DNA from the insertion site of the element from line 02382 (Figure 5A).
Figure 5.—
Deletion analysis of piggyBac-neighboring DNA. The piggyBac elements in lines 02382 (A), 04432 (B), and 06451 (C) were relocated to new genomic positions (shown in parentheses) with progressively less neighboring chromosomal DNA, beginning with 1 kb of neighboring DNA. The mean proportion (± SD; n = 3) of flies in a remobilization testcross with mosaic eye color is shown. A one-way ANOVA was performed to compare the effect of the amount of element-flanking DNA on element activity. “piggyBac-0bp” refers to a piggyBac element relocated in the same way to the positions indicated but without any of the original neighboring chromosoml DNA. There was a significant effect of the amount of element-flanking DNA on element activity: F[7,16]=875.32, P < 0.0001 (A ); F[4,10]=472.18, P < 0.0001 (B ); and F[5,12]=341.47, P < 0.0001 (C). Post-hoc comparisons of all pairs of means using Tukey's HSD test indicated which elements had significantly different activities (P < 0.05). Elements with significantly different means are indicated with different lower case letters.
The deletion experiment described above was repeated, in part, using the elements from lines 04432 and 06451, which also have low rates of remobilization when in their original genomic locations. Both elements were relocated to position 86Fb along with 200 bp of flanking chromosomal DNA. Like the element from line 02382, the element from line 04432 had a significantly higher activity when it was relocated with only 200 bp of its original flanking chromosomal DNA (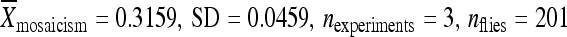 ) compared to the element in its original genomic location (
) compared to the element in its original genomic location (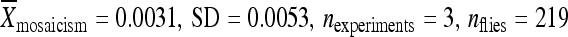 ) and at 86Fb along with 1 kb of flanking genomic DNA (
) and at 86Fb along with 1 kb of flanking genomic DNA (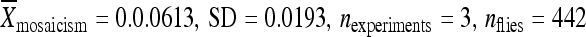 ) (Figure 5B). The element from line 06451, when relocated to position 86Fb with 200 bp of its original flanking chromosomal DNA, had a somewhat higher remobilization activity (
) (Figure 5B). The element from line 06451, when relocated to position 86Fb with 200 bp of its original flanking chromosomal DNA, had a somewhat higher remobilization activity (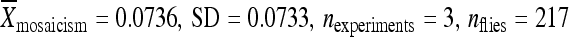 ) compared to the element in its original genomic location (
) compared to the element in its original genomic location (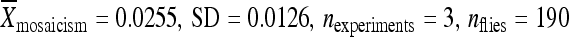 ) and relocated with 1 kb of flanking chromomal DNA (at 51D:
) and relocated with 1 kb of flanking chromomal DNA (at 51D: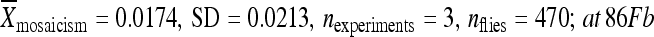 :
: 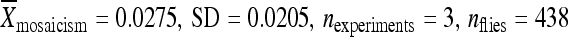 ), but these differences were not statistically significant (P > 0.05) (Figure 5C).
), but these differences were not statistically significant (P > 0.05) (Figure 5C).
Position effects are portable to plasmids:
To test whether the intrinsic local properties of piggyBac-flanking chromosomal DNA had similar effects on element mobility when elements were residing on plasmids, two elements with widely differing activity levels as a result of their local chromosomal context (low-activity element in line 02382 and high-activity element in line 05577) were cloned into plasmids along with 1 kb of flanking chromosomal DNA and used as gene vectors to create transgenic D. melanogaster using routine germline transformation methods. For comparison, each experimental vector was co-injected with a commonly used piggyBac vector with the Enhanced Green Fluorescent Protein gene under the regulatory control of a central nervous system-specific promoter (3XP3) (Horn and Wimmer 2000). As expected, after analyzing the germlines of 355 and 292 fertile G0 adults arising from embryos injected with high-and low-activity elements, respectively, along with the reference vector, we observed no significant difference in the frequency of transformation with the reference vector (χ2=3.5844, P = 0.0583) (Table 2). However, the two experimental vectors had distinctly different activities. The high-activity vector resulted in ∼ 10-fold more transformed germlines than the low-activity vector, showing that the elements retained the same relative activity levels as when the elements were integrated into chromosomes (0.121 [05577] ± 0.033 [95% C. I.]; 0.014[02382] ± [95% C. I.]; χ2=90.0775, P = 0.0000) (Table 2).
TABLE 2.
Flanking DNA affects piggyBac elements on plasmids
| Vector mixturea | Fertile G0 germlines tested | germlines producing w+ progeny | germlines producing DsRed progeny |
|---|---|---|---|
| 02382∷1000b + PB{3xP3DsRed} | 292 | 4 | 42 |
| 05577∷1000b + PB{3xP3DsRed} | 355 | 43 | 77 |
| χ2 | 90.0775 | 3.5844 | |
| P | 0 | 0.05832 |
A mixture of two vectors and a transposase-expressing plasmid. Each plasmid is at 0.25 mg/ml.
The piggyBac elements in these vectors were flanked by 1 kb of their original chromosomal insertion site DNA.
piggyBac elements flanked by nuclear polyhedrosis virus sequences are suboptimal:
We have shown that the local genomic neighborhood influences the activity of piggyBac elements whether they are integrated within chromosomes of D. melanogaster or within plasmids. Most, if not all, piggyBac vectors in use currently are derived from the original IFP2 transposable element isolated from the FP locus of a nuclear polyhedrosis virus following passage through Tricoplusia ni cells (Cary et al. 1989). These piggyBac gene vectors are flanked on each end by ∼ 150 bp of nuclear polyhedrosis virus DNA. In light of the profound effects flanking DNA can have on element mobility, we tested the remobilization activity of the piggyBac element on the gene vector PB{RB} (Thibault et al. 2004), which was used to create the transgenic lines used in this study, by cloning the element along with 1 kb of the flanking plasmid DNA, including the 150 bp of nuclear polyhedrosis virus DNA, and integrated the plasmid into cytological position 86Fb. The activity of this element was measured by determining the frequency of eye-color mosaicism in the presence of the standard source of piggyBac transposase used in this project. This common form of piggyBac showed moderate-to-high levels of activity (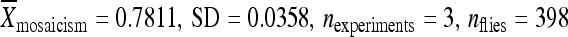 ); however, 5 of the 19 piggyBac elements tested in this study had flanking DNA sequences that resulted in significantly higher levels of piggyBac activity, indicating that PB{RB}, and probably all piggyBac vectors currently in use, are suboptimal, at least for use in D. melanogaster as a consequence of the influence of the DNA sequences flanking the element.
); however, 5 of the 19 piggyBac elements tested in this study had flanking DNA sequences that resulted in significantly higher levels of piggyBac activity, indicating that PB{RB}, and probably all piggyBac vectors currently in use, are suboptimal, at least for use in D. melanogaster as a consequence of the influence of the DNA sequences flanking the element.
DNA bendability is associated with position-dependent piggyBac activity:
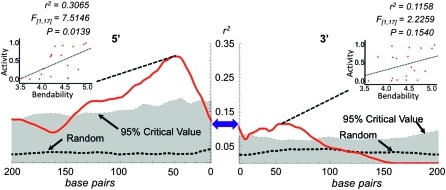
Because local bending of transposable element DNA is known to be important during some transposition reactions and because DNA-bending proteins are also known to be involved in the transposition of some transposable elements, we tested, using linear regression modeling, the association between predicted bendability of DNA flanking the piggyBac element and element activity (Makris et al. 1990; van Gent et al. 1997; Sewitz et al. 2003; Zayed et al. 2003; Wardle et al. 2005). Because we did not know how much of the flanking DNA sequence to consider when calculating average bendability, we considered all lengths ranging from 1 to 200 bp from the terminal inverted repeats and the TTAA target-site duplication. For a given length of flanking DNA, the average bendability was calculated for each element (see materials and methods) and, in conjunction with the activity measurements for each element, Pearson's coefficient of determination, r2, was calculated (Figure 6). As a control, 25 sets of random sequences were associated with each element-activity measurement and r2 was calculated considering the average bendability of all lengths of randomized flanking DNA from 1 to 200 bp. Prior to constructing the data sets with randomized sequences, the nucleotide content was calculated over 1 kb of DNA flanking the elements in this analysis. The resulting nucleotide abundance table was used to construct the randomized sequences. Thus the average nucleotide abundance of the randomized DNA sequences was the same as the experimental data. From the randomized data sets, the means and 95% critical values of Pearson's coefficients of determination were calculated and plotted (Figure 6). A significant positive association was seen between the activity of elements and the predicted bendability of the sequences flanking the 5′ terminal inverted repeat of the piggyBac element. This association was maximal when the predicted bendability of the first ∼ 50 bp were considered (r2=0.3065, F[1,17]=7.5146, P = 0.0139). A similar trend in the association between the activity of elements and the predicted bendability of approximately the first 50 bp flanking the 3′ terminal inverted repeat of the piggyBac element (r2 = 0.1158, F[1,17] = 2.2259, P = 0.1540), although the association was much weaker than that seen at the 5′ end and was not statistically significant. A multiple linear regression analysis using the predicted bendability of the first ∼ 50 bp of DNA flanking the 5′ and 3′ terminal inverted repeats of piggyBac as predictor variables accounted for 60.34% of the variance observed among the elements at different genomic positions (r2adjusted=0.6034).
Figure 6.—
Association between predicted bendability of piggyBac-neighboring DNA and element activity. The association between the activity of piggyBac elements and the bendability (predicted) of all lengths of piggyBac-neighboring DNA up to 200 bp from the terminal inverted repeats was calculated using linear regression analysis. The coefficient of determination, r2, was calculated and plotted against the length of DNA used in the analysis (red line). A set of 25 random sequences was generated as described in materials and methods, and the association between piggyBac activity and predicted bendability of the randomized flanking DNA was determined for each of the data sets. The mean coefficient of determination (r2) was plotted (dashed line; n = 25) along with the 95% critical value (gray shaded area). The strength of the association depended upon how much neighboring DNA was considered in the analysis. Where r2 is maximal, the bendability and activity data are shown with the results of the linear regression analysis (inset graphs). The location of the flanking DNA relative to the piggyBac element (blue double arrow) is indicated by “5′” and “3′.”
DNA bendability is not associated with the target-potential of sites:
To begin to examine the relationship between the characteristics of sites that determine the propensity of piggyBac elements to excise/transpose and the characteristics that determine whether they will be preferred targets for piggyBac integration, 460 “hot spots” for piggyBac integration in the genome of D. melanogaster were analyzed in a manner similar to that described above. Sites in which piggyBac integrations have been reported two or more times in exactly the same site and orientation were selected and the average bendability of the first 50 bp flanking the 5′ terminal-inverted-repeat was calculated as described (see Materials And Methods). Using a linear model, Pearson's coefficient of determination, r2, was calculated and found to be insignificantly different from zero (r2 = 0.0003, F(1,458) = 0.1336, P = 0.7149) revealing no association between the “hotness” of a site (number of independent integrations) and the predicted bendability of the region of DNA found to be associated with remobilization activity.
DISCUSSION
We have shown that the location of piggyBac transposable elements within the genome of D. melanogaster is a major determinant of their activities and have begun to elucidate how this position effect occurs. In this study we used a somatic remobilization assay that estimated element excision and/or transposition activity by measuring the frequency of eye-color mosaicism in transgenic lines containing a single mini-white gene-containing piggyBac element in the presence of a constant source of piggyBac transposase. Because each eye of adult D. melanogaster is composed of ∼ 800 ommatidia, even excision and/or transposition events that resulted in a qualitative change in pigmentation in a small number of ommatidia (i.e., resulting from a remobilization event that took place late in the development of the eye) could be detected, making eye-color mosaicism a sensitive means for measuring piggyBac movement. The least active element in this study was in line 04432 with a frequency of mosaicism of only 0.3% while the most active element was in line 05577 with a frequency of mosaicism of 99.5%. This large difference in activity is significant because essentially all of the variance in element activity measured in this experiment can be attributed to the difference in the genomic position of the elements. All piggyBac elements analyzed in this study were identical with the exception of the elements in lines 02382 and 03565, which differed only slightly from the other elements (Thibault et al. 2004). Furthermore, we estimate that the difference in activities of the elements in lines 04432 and 05577 to be 100-fold although we think that this is probably an underestimate. Flies were scored as having mosaic eyes if they had a single detectable patch of ommatidia of any size with pigmentation different from the parental line. In this experiment, no attempt was made to count the number of clones within mosaic eyes, which varied among the lines. For example, line 05577 displayed not only a high frequency of mosaicism but also evidence of multiple remobilizations in those eyes that were mosaic—numerous clones of ommatidia with differing levels of pigmentation resulting from element excision or transposition. The difference in element activity between the elements in lines 05577 and 04432 therefore exceeded two orders of magnitude. While elements 05577 and 04432 represented the extremes recorded in this experiment, they were not exceptional. Four other elements had high levels of activity that were not resolvable statistically from 05577, and five other elements had levels of activity similar to 04432 (Figure 1). These data show that, in the presence of functional piggyBac transposase, position is a major, or perhaps the major, determinant of piggyBac element activity in D. melanogaster.
Others have reported differences in transposable element activity as a function of an element's position within a genome, but this is the first study to tightly control for other factors that could also affect mobility, including differences in element sequence, content and structure, and genetic background. For example, Berg and Spradling (1991) reported a sixfold difference in the germline transposition activity of two identical P elements in different locations on the X chromosome. While Bancroft and Dean (1993) reported that Ds elements in different positions in the genome of A. thaliana had varying amounts of somatic and germline activity. Smith et al. (1996) found no significant difference in the activity of 5 Ds elements inserted at different locations within A. thaliana (Bancroft and Dean 1993; Smith et al. 1996). The excision and transposition rates of MuDR-1 elements in maize are also sensitive to chromosomal position (Lisch et al. 1995). The magnitude of the differences among position-dependent remobilization rate reported here is particularly notable in light of this earlier work and that essentially all of the observed variance in activity could be attributed to position.
The influence of neighboring DNA has been reported in a number of other recombination reactions. Vigdal et al. (2002) tested the effects of altering the sequence of DNA directly flanking the terminal inverted repeats of active Sleeping Beauty elements on element excision activity. They altered only the sequence of the first six base pairs adjacent to the TA target-site duplication, and of the 17 sequence variations tested, only 2 resulted in significantly different excision rates of the Sleeping Beauty element, leading the authors to conclude that sequences directly flanking excision sites have no significant effect on Sleeping Beauty transposition (Vigdal et al. 2002). In contrast to this, Cuomo et al. (1996) reported that recombination involving the transposable element-like recombination signal sequences (RSS) during immunoglobulin and T-cell receptor gene assembly was sensitive to changes in the flanking sequence (Cuomo et al. 1996). Changes involving only the first few nucleotides of flanking DNA in coding sequences immediately adjacent to the heptamer had large effects on the efficiency of recombination in vitro. Recombination frequencies could be reduced as much a 1000-fold when a polythymidylate tract flanked the RSS (Cuomo et al. 1996). The authors speculated that RAG protein-mediated distortion involving the flanking DNA was necessary for subsequent trans-esterification reactions, and this could account for the sensitivity of this recombination reaction to sequences outside the RSS.
Liu et al. (2006) found that the removal of integration host factor and the unfolding of the Tn10 transpososome during the transposition reaction was dependent upon the presence of terminal-inverted-repeat-flanking DNA. Indeed, they suggested that the energy needed to drive transposition comes in part from the distorted DNA sequences flanking the ends of the element (Liu et al. 2006). More recently, Mizuuchi et al. (2007) found that DNA flanking the phage Mu and its configuration played a critical role in determining the enzymatic activity of Mu transposase. Previous foot-printing experiments demonstrated that the transpososome included flanking DNA (Lavoie et al. 1991; Mizuuchi and Adzuma 1991; Crellin and Chalmers 2001) and Mizuuchi et al. (2007) suggest that the configuration of this flanking DNA could influence the reactions taking place in the catalytic core (Mizuuchi et al. 2007).
While these systems illustrate how transposable element-flanking DNA might be important to transposition/excision reactions, they highlight the role of a relatively small number of base pairs immediately adjacent to the elements. The results reported here suggest that a much more extensive region of flanking DNA extending up to perhaps 500 bp from the terminal inverted repeat can significantly influence element activity in vivo. This suggests that the effects seen here are somewhat distinct from those previously described, and it is not yet clear whether the influence of flanking DNA reported here reflects a direct or indirect role of these sequences in excision/transposition reactions. Cell-free transposition reactions will permit the influence of the flanking DNA sequences to be compared. It is likely that the flanking DNA influences the local chromatin structure at the terminal regions of the transposable element and affects the accessibility of transposase to critical binding sites and substrates. This is consistent with the observation that predicted DNA bendability of element-flanking DNA can account for ∼ 60% of the variance in element activity observed in this experiment. The trinucleotide bendability estimates used to estimate DNA bendability in this experiment were derived from empirical studies involving sensitivity to DNAse I and nucleosome protection studies (Brukner et al. 1995; Gabrielian and Pongor 1996). The results reported here may be a reflection of how nucleosome positioning may be a critical determinant of element activity.
The findings reported in this study have at least two additional implications. First, although it is not clear whether other class II transposable elements will show the same range of effects of genomic position on remobilization potential, these position effects will need to be considered carefully when interpreting distributions of elements within genomes and the degree to which this reflects aspects of the element's integration site preferences. There have been numerous studies that describe the distributions of various DNA transposons within genomes, and without exception the distributions are found to be non-uniform. The patterns of element distribution vary among elements, with each having characteristic regional and intragenic preferences. For example, Thibault et al. (2004) generated 18,225 piggyBac insertions in D. melanogaster and noted hot spots and cold spots for element accumulation, which were interpreted as reflecting preferences for certain integration sites. While elements do have integration site preferences as reflected in target-site duplications associated with element integration, the patterns of element accumulation within genomes are a reflection not only of integration site preferences but also of patterns of remobilization potential, which, as we show in this study, are determined by intrinsic properties of local regions of the genome. The patterns of Class II transposable elements that undergo both excision and transposition within genomes will be a function of “in rates” and “out rates”, but when interpreting element distribution data, “out rates” have been implicitly assumed to be either insignificantly low or uniform across the genome. Based on the data presented here it seems clear that the “out rates” of piggyBac elements in D. melanogaster are not uniform across the genome. At the moment, the relationship between a site's potential as a target for integration and its potential to promote or condition subsequent remobilization of integrated elements is not known. If those two characteristics of sites are independent, then it is possible that two sites within a genome that are equally preferred as integration sites may show unequal frequencies of site occupancy because of differences in the effects of neighboring DNA on element remobilization. Likewise, sites with equal site-occupancy frequencies may have different target site potentials. Our data highlight the importance of understanding the characteristics of sites that favor integration and remobilization and whether there is any relationship among them. We were unable to find any data in the published literature that addressed this question although Bender and Kleckner reported unpublished observations that Tn10 elements residing in sites known to be preferred integration sites excised at the same rate as elements in “cold” sites, suggesting that what determines a site's target potential is independent of what determines its remobilization potential (1992). Because our data revealed an association (r2 ≈ 0.3) between element remobilization and the predicted bendability of the first 50 bp of DNA flanking the 5′ terminal-inverted-repeat of piggyBac we determined if this characteristic was also associated with the targetability of sites. After identifying 460 piggyBac insertion sites in D. melanogaster where two or more elements had been reported in the same orientation, we found no association (r2 ≈ 0.0) between local predicted bendability and target “hotness”. This observation is consistent with those of Bender and Kleckner (1992), but stronger tests of the hypothesis that site-characteristics conditioning remobilization are independent of those conditioning integration are clearly needed.
Our results also have important implications for the design and assembly of piggyBac-based gene vectors and potentially other class II transposable element-based gene vectors. The strong influence of neighboring DNA sequences on excision and transposition activity of piggyBac was retained when elements and their flanking DNA were relocated to new genomic locations and also when the elements and their flanking DNA sequences were placed on plasmids. The relative activities of the elements in lines 05577 and 02382 were retained when the elements and 1 kb of flanking genomic DNA were moved onto plasmids and used as germline transformation vectors in D. melanogaster. Therefore, piggyBac elements can have widely different activities as gene vectors because of the strong influence of flanking DNA sequences. Knowing the “rules” that govern this neighborhood effect will permit gene vectors that have significantly more activity to be assembled without necessarily modifying the original element. In combination with element and transposase modifications, optimizing the sequence context in which an element resides could result in large increases in element activity. Essentially, all piggyBac vectors currently in use have been derived from the original IFP2 transposable element isolated from the FP locus of a nuclear polyhedrosis virus following passage through Tricoplusia ni cells (Cary et al. 1989). These piggyBac gene vectors are flanked on each end by ∼ 150 bp of nuclear polyhedrosis virus DNA, and while these sequences are conducive to high remobilization activity in D. melanogaster, as shown here, they are far from optimal in this species.
While we have identified DNA sequence contexts that are conducive to active piggyBac excision/transposition in D. melanogaster, we do not know if the same sequence contexts will be conducive to piggyBac movement in other species. This is an important question because, clearly, gene vectors are currently exchanged and used interspecifically without consideration of the possibility that functionality could be affected by species-specific “context effects.” Species-specific differences in element activities have typically been interpreted as reflecting differences in transposase activities or other uncontrollable host factors. We have made preliminary observations showing that piggyBac elements and their flanking chromosomal DNA from the yellow fever mosquito, Aedes aegypti, where these elements do not remobilize (Sethuraman et al. 2007), when relocated to the genome of D. melanogaster (position 86Fb, a site used to host relocated piggyBac elements in this study) can remobilize at high rates (Azhahianambi Palavasam, unpublished observation). These observations are consistent with the idea that local sequence “context-effects” will be species-specific.
Finally, while the data presented here inform us of piggyBac's sensitivity to DNA sequence context, they do not allow us to draw conclusions about other elements. Placing other transposable elements, e.g. P-elements, Hermes, in DNA sequence contexts with defined characteristics based on experiments with piggyBac will permit the generality of our results to be determined. These experiments are ongoing.
Acknowledgments
Robert Alford provided us with valuable discussion and suggestions during the course of this study. This work was supported by grant AI70812 from the National Institutes of Health National Institute of Allergy and Infectious Diseases.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.122168/DC1.
Available freely online through the author-supported open access option.
References
- Bancroft, I., and C. Dean, 1993. Factors affecting the excision frequency of the maize transposable element Ds in Arabidopsis thaliana. Mol. Gen. Genet. 240 65–72. [DOI] [PubMed] [Google Scholar]
- Bender, J., and N. Kleckner, 1992. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc. Natl. Acad. Sci. USA 89 7996–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, C. A., and A. C. Spradling, 1991. Studies on the rate and site-specificity of P element transposition. Genetics 127 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof, J., R. K. Maeda, M. Hediger, F. Karch and K. Basler, 2007. An optimized transgenesis system for Drosophila using germ-line-specific phi C31 integrases. Proc. Natl. Acad. Sci. USA 104 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouuaert, C. C., and R. M. Chalmers, 2010. Gene therapy vectors: the prospects and potentials of the cut-and-paste transposons. Genetica 138 473–484. [DOI] [PubMed] [Google Scholar]
- Brukner, I., R. Sanchez, D. Suck and S. Pongor, 1995. Sequence-dependent bending propensity of DNA as revealed by DNase-I-parameters for trinucleotides. EMBO J. 14 1812–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, D. F., and S. C. Fahrenkrug, 2010. Using transposons to increase the efficiency of embryo and cellular transgenesis. Transgenic Res. 19 132–133. [Google Scholar]
- Cary, L. C., M. Goebel, B. G. Corsaro, H. G. Wang, E. Rosen et al., 1989. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172 156–169. [DOI] [PubMed] [Google Scholar]
- Crellin, P., and R. Chalmers, 2001. Protein-DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. EMBO J. 20 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo, C. A., C. L. Mundy and M. A. Oettinger, 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. R., 1989. P elements in Drosophila melanogaster, pp. 439–484 in Mobile DNA, edited by D. E. Berg and M. M. How. American Society for Microbiology, Washington, DC.
- Feschotte, C., and E. J. Pritham, 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41 331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, M. P., A. C. Groth, M. P. Calos and R. Nusse, 2007. Creating transgenic Drosophila by microinjecting the site-specific phi C31 integrase mRNA and a transgene-containing donor plasmid. Nat. Protoc. 2 2325–2331. [DOI] [PubMed] [Google Scholar]
- Gabrielian, A., and S. Pongor, 1996. Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Lett. 393 65–68. [DOI] [PubMed] [Google Scholar]
- Groth, A. C., M. Fish, R. Nusse and M. P. Calos, 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phi C31. Genetics 166 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler, A. M., and R. A. Harrell, 1999. Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol. Biol. 4 449–458. [DOI] [PubMed] [Google Scholar]
- Horn, C., and E. A. Wimmer, 2000. A versatile vector set for animal transgenesis. Dev. Genes Evol. 210 630–637. [DOI] [PubMed] [Google Scholar]
- Ivics, Z., and Z. Izsvak, 2006. Transposons for gene therapy! Curr. Gene Ther. 6 593–607. [DOI] [PubMed] [Google Scholar]
- Ivics, Z., M. A. Li, L. Mates, J. D. Boeke, A. Nagy et al., 2009. Transposon-mediated genome manipulation in vertebrates. Nature Methods 6 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G., 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115 49–63. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., and D. R. Lisch, 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55 1–24. [DOI] [PubMed] [Google Scholar]
- Kitamura, K., S. Hashida, T. Mikami and Y. Kishima, 2001. Position effect of the excision frequency of the Antirrhinum transposon Tam3: implications for the degree of position-dependent methylation in the ends of the element. Plant Mol. Biol. 47 475–490. [DOI] [PubMed] [Google Scholar]
- Lavoie, B. D., B. S. Chan, R. G. Allison and G. Chaconas, 1991. Structural aspects of a higher-order nucleoprotein complex: induction of an altered DNA-structure at the Mu-host junction of the Mu-type-1 transpososome. EMBO J. 10 3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis, R., T. Hazelrigg and G. M. Rubin, 1985. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science 229 558–561. [DOI] [PubMed] [Google Scholar]
- Liao, G., E. J. Rehm and G. M. Rubin, 2000. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97 3347–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch, D., P. Chomet and M. Freeling, 1995. Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics 139 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. X., S. Sewitz, P. Crellin and R. Chalmers, 2006. Functional coupling between the two active sites during Tn10 transposition buffers the mutation of sequences critical for DNA hairpin processing. Mol. Microbiol. 62 1522–1533. [DOI] [PubMed] [Google Scholar]
- Liu, S. Z., C. T. Yeh, T. M. Ji, K. Ying, H. Y. Wu et al., 2009. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PloS Genet. 5 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris, J. C., P. L. Nordmann and W. S. Reznikoff, 1990. Integration host factor plays a role in IS50 and Tn5 transposition. J. Bacteriol. 172 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, C. D., and G. J. Hannon, 2009. Small RNAs as guardians of the genome. Cell 136 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi, K., and K. Adzuma, 1991. Inversion of the phosphate chirality at the target site of Mu-DNA strand transfer: evidence for a one-step transesterification mechanism. Cell 66 129–140. [DOI] [PubMed] [Google Scholar]
- Mizuuchi, M., P. A. Rice, S. J. Wardle, D. B. Haniford and K. Mizuuchi, 2007. Control of transposase activity within a transpososome by the configuration of the flanking DNA segment of the transposon. Proc. Natl. Acad. Sci. USA 104 14622–14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. P., and H. E. Varmus, 1994. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 13 4704–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Lopez, M., and J. L. Garcia-Perez, 2010. DNA transposons: nature and applications in genomics. Curr. Genomics 11 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard, D. J., K. H. J. Gordon, A. H. Buck and F. M. Jiggins, 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. B Biol. Sci. 364 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham, E. J., 2009. Transposable elements and factors influencing their success in eukaryotes. J. Hered. 100 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss, D., F. D. Bushman and A. P. Wolffe, 1994. Human-immunodeficiency-virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA 91 5913–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak, P. M., and H. E. Varmus, 1992. Nucleosomes, DNA-binding proteins, and DNA-sequence modulate retroviral integration target site selection. Cell 69 769–780. [DOI] [PubMed] [Google Scholar]
- Rebollo, R., B. Horard, B. Hubert and C. Vieira, 2010. Jumping genes and epigenetics: towards new species. Gene 454 1–7. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2010 The R Project for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnsonschlitz, W. K. Benz et al., 1988. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics 118 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T. D., and K. J. Livak, 2008. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3 1101–1108. [DOI] [PubMed] [Google Scholar]
- Sethuraman, N., M. J. Fraser, P. Eggleston and D. A. O'Brochta, 2007. Post-integration stability of piggy Bac in Aedes aegypti. Insect Biochem. Mol. Biol. 37 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewitz, S., P. Crellin and R. Chalmers, 2003. The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. Nucleic Acids Res. 31 5868–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin, R. K., and R. Martienssen, 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8 272–285. [DOI] [PubMed] [Google Scholar]
- Smith, D., Y. Yanai, Y. G. Liu, S. Ishiguro, K. Okada et al., 1996. Characterization and mapping of Ds-GUS-T-DNA lines for targeted insertional mutagenesis. Plant J. 10 721–732. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. Dompe et al., 2004. P and piggyBac transposons display a complementary insertion spectrum in Drosophila: a multifunctional toolkit to manipulate an insect genome. Nat. Genet. 36 283–287. [DOI] [PubMed] [Google Scholar]
- Tweedie, S., M. Ashburner, K. Falls, P. Leyland, P. McQuilton et al., 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acid Res. 37 D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenDriessche, T., Z. Ivics, Z. Izsvak and M. K. L. Chuah, 2009. Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood 114 1461–1468. [DOI] [PubMed] [Google Scholar]
- van Gent, D. C., K. Hiom, T. T. Paull and M. Gellert, 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16 2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdal, T. J., C. D. Kaufman, Z. Izsvak, D. F. Voytas and Z. Ivics, 2002. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323 441–452. [DOI] [PubMed] [Google Scholar]
- Wardle, S. J., M. O'Carroll, K. M. Derbyshire and D. B. Haniford, 2005. The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev. 19 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed, H., Z. Izsvak, D. Khare, U. Heinemann and Z. I, 2003. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31 2313–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]