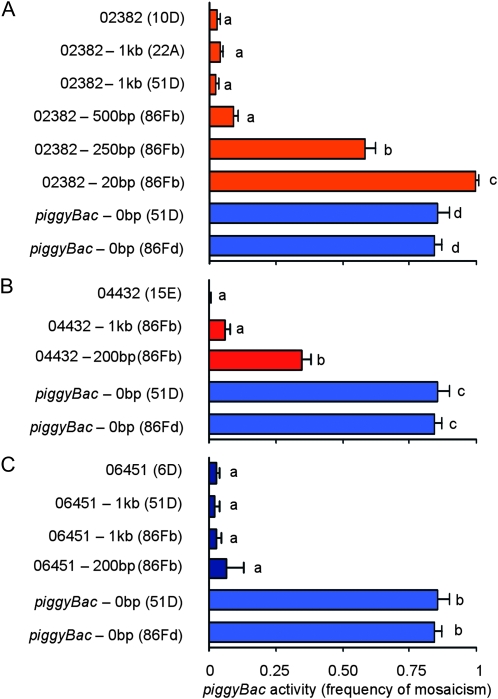
Figure 5.—
Deletion analysis of piggyBac-neighboring DNA. The piggyBac elements in lines 02382 (A), 04432 (B), and 06451 (C) were relocated to new genomic positions (shown in parentheses) with progressively less neighboring chromosomal DNA, beginning with 1 kb of neighboring DNA. The mean proportion (± SD; n = 3) of flies in a remobilization testcross with mosaic eye color is shown. A one-way ANOVA was performed to compare the effect of the amount of element-flanking DNA on element activity. “piggyBac-0bp” refers to a piggyBac element relocated in the same way to the positions indicated but without any of the original neighboring chromosoml DNA. There was a significant effect of the amount of element-flanking DNA on element activity: F[7,16]=875.32, P < 0.0001 (A ); F[4,10]=472.18, P < 0.0001 (B ); and F[5,12]=341.47, P < 0.0001 (C). Post-hoc comparisons of all pairs of means using Tukey's HSD test indicated which elements had significantly different activities (P < 0.05). Elements with significantly different means are indicated with different lower case letters.