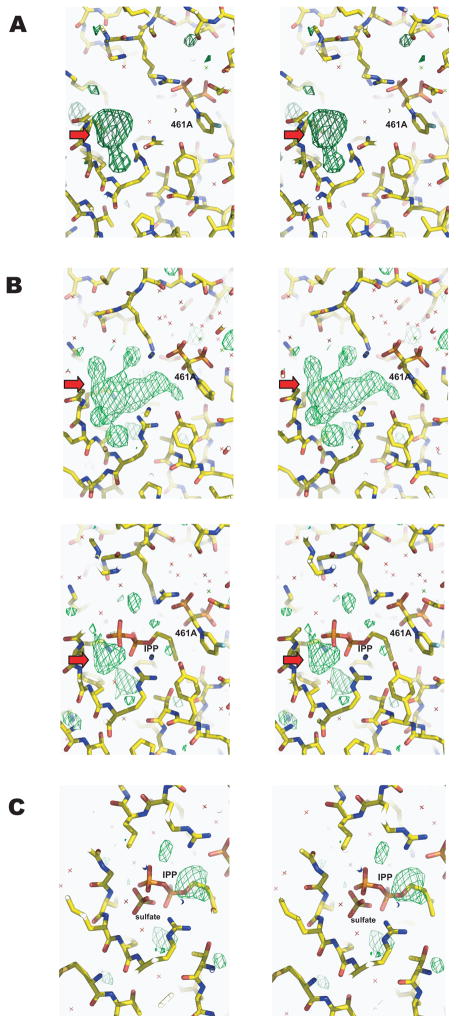
Figure 4. Comparison of electron density maps at the homoallylic site of TcFPPS/461A and TcFPPS/461A/IPP complexes showing the presence of a sulfate molecule.
Fo-Fc maps are shown as green meshes. In (A), electron density maps of TcFPPS/461A complex (no IPP) show extra densities (red arrows) in the Fo-Fc OMIT map. This density corresponds to a sulfate molecule at the homoallylic site in the apo enzyme structure9. For the TcFPPS/461A/IPP complex, the Fo-Fc OMIT map (B, upper panel) show much bigger densities than those in the TcFPPS/461A complex, indicating possible occupancy by IPP. However, an IPP molecule cannot fully account for those densities and leaves residual density (red arrows in B, lower panel) at a location corresponding to the density for the sulfate molecule in the TcFPPS/461A complex (A). When both IPP and sulfate are built into the homoallylic site of the TcFPPS/461A/IPP complex with 50% occupancy each (C), the model fits the electron density map well except for some residual density at the isopentenyl end of IPP, indicating disordering of this region. All Fo-Fc maps are contoured 2.8 σ.