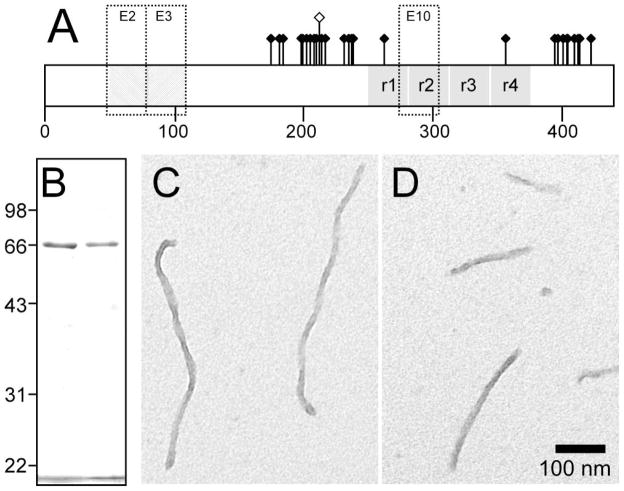
Fig. 1. Comparison of 2N4R and 2N4R-T212E tau preparations.
(A) Distribution of ~30 hydroxyamino acid residues affected by phosphorylation depicted on isoform 2N4R. This isoform contains alternatively spliced exons 2 and 3 (E2 and E3), each of which encodes an acidic 29-residue segment, and exon 10 (E10), which encodes an additional microtubule binding repeat. Pseudophosphorylation mutant T212E is distinguished graphically by a raised hollow symbol. (B) Comparison of recombinant 2N4R (left lane) and 2N4R-T212E (right lane) protein preparations by SDS-polyacrylamide gel electrophoresis (10% acrylamide) and Coomassie blue staining. The bands migrating at ~66 kDa correspond to these tau proteins [33], whereas the band below 22 kDa corresponds to the dye front. The preparations were consistent with respect to protein concentration and purity. (C,D) Morphology of synthetic tau filaments. Full-length wild-type 2N4R tau (C) and mutant 2N4R-T212E (D) were incubated (1 μM concentration) without agitation in the presence of 100 μM Thiazine red (24 h at 37°C) and viewed by transmission electron microscopy. 2N4R-T212E produced unbranched filaments ~16 nm in diameter with no obvious differences in morphology or length distribution from wild-type 2N4R. Scale bar = 100 nm.